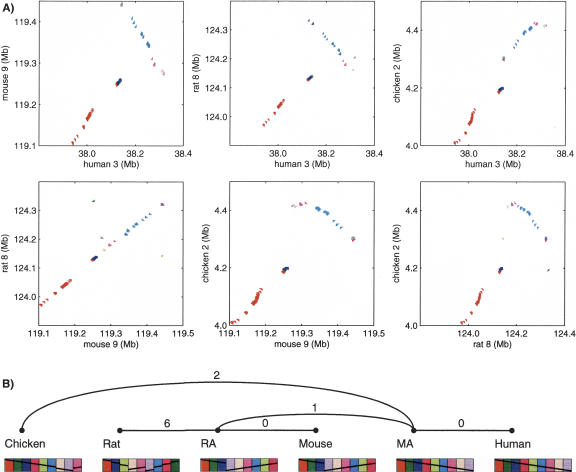
Figure 5.
(A) Dot-plots of similarity anchors in a four-way synteny block with microrearrangements. The block was computed by GRIMM-Synteny on sequence-based data in run 300K. The block length is 380 kb in human, 340 kb in mouse, 364 kb in rat, and 419 kb in chicken. The anchors divide into nine groups, indicated by different colors; these groups have a different order and orientation in each genome. Within each group, the anchors have the same relative order in all genomes (or an overall flip of the whole group), but their absolute spacing is not preserved. (B) Microrearrangement history for the same block as generated by MGR. The nine colored blocks correspond to the nine groups of anchors in the dot-plots. The minimum number of inversions required to convert between synteny blocks in two genomes is shown on each edge of the tree.