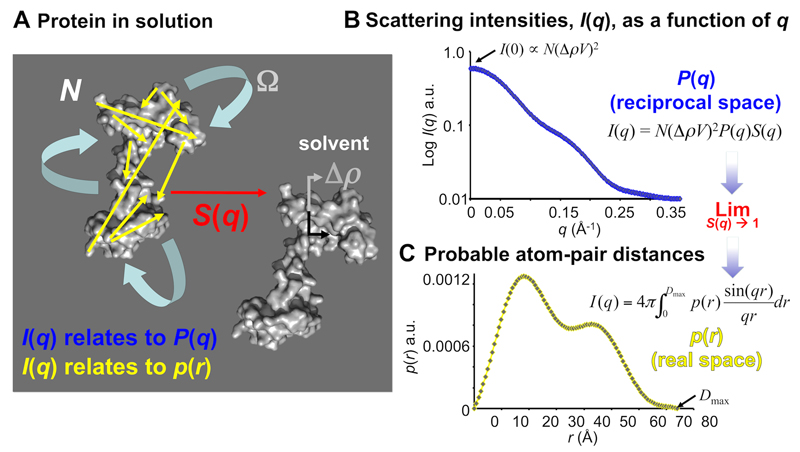
Figure 1. Scattering basics.
A. Macromolecules in solution, e.g., proteins (represented as grey blobs), undergo rotational and translational motion and experience long-range interactions with neighbouring particles. The SAS intensities measured from an isotropically tumbling (Ω) monodisperse sample are dependent on a number of factors, of which the form factor, P(q), is of most interest to structural biologists. It is from P(q) that structural parameters and low resolution models of the macromolecules can be obtained. The form factor of the scattering intensities in reciprocal space relate to the real space distribution, p(r), of all time-preserved, i.e., correlated, pair-distances between scattering centres of the molecule (yellow arrows). In the small-angle regime, these correlated distances are otherwise absent in the solvent. However, as all atoms can scatter radiation, solvent scattering contributions have to be accurately subtracted from the sample scattering to reveal P(q) from the macromolecules. The magnitude of the intensities will then depend on: i) the number of particles in a sample (N); ii) the volume squared of the macromolecule (V2); iii) the difference in scattering length density, or the contrast, squared against the solvent (Δρ2) and; iv) scattering arising from correlated distances of closest approach between particles (interparticle interference, or structure factors, S(q)). The purity, concentration, contrast and how well a solvent is matched to a sample can be directly controlled during sample preparation. B. SAS data are usually collected on 2D detectors and radially averaged to produce 1D profiles of scattering intensity, I(q), as a function of angle, q. After solvent subtraction, I(q) vs q encodes P(q) from each-and-every macromolecule in a sample weighted by N(ΔρV)2 and S(q). Longer distance separations are represented at lower angles and vice-versa. At zero angle, I(0), the magnitude of the scattering is proportionate to the total volume squared and concentration of the macromolecules. C. If S(q) limits to 1, i.e., when the system is infinitely dilute and interparticle effects are absent, modelling the indirect inverse Fourier transform of I(q) vs q produces the real-space p(r) vs r from which the radius of gyration, Rg, maximum particle dimension, Dmax, and low resolution particle shape and structure can be determined.