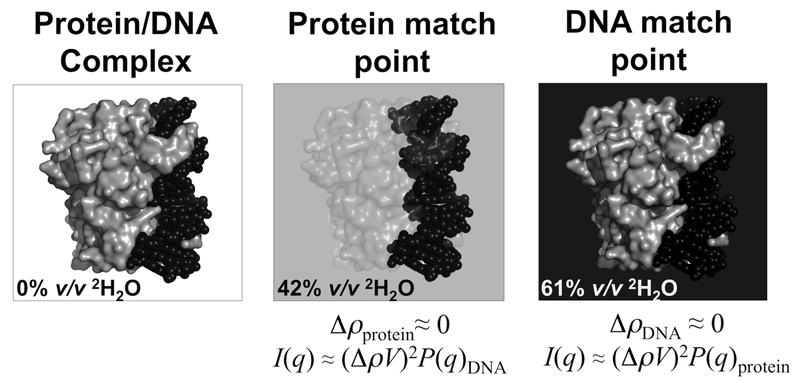
Figure 4. Principle of contrast matching.
If a macromolecular complex consists of individual components that have different average scattering length densities, it is possible to match out the scattering contributions of a component by placing the complex in a solvent with the same average scattering length density as that component. Illustrated here is a protein/DNA complex (grey surface and black spheres, respectively). For example, using neutrons, if the complex is placed into ~42% v/v 2H2O, i.e., the protein match point, the measured coherent scattering of the SANS profile will be dominated by the DNA, from which the disposition of the DNA in the complex can be determined. Raising the % v/v 2H2O to 61% matches out the DNA scattering contribution so that the SANS profile is dominated by coherent scattering from the protein.