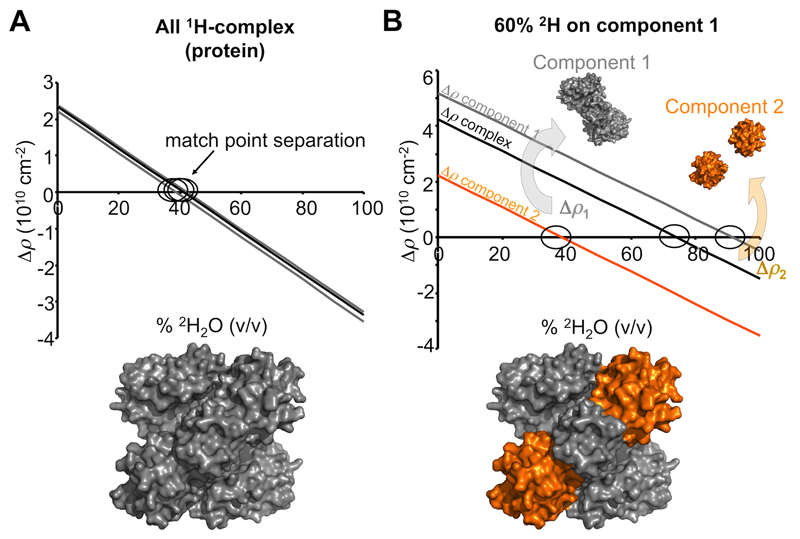
Figure 5. The effect of non-exchangeable deuterium labelling a component for SANS with contrast variation experiments.
A. As there is very little difference in the average 1H per unit volume for proteins, the neutron contrasts calculated at different % v/v 2H2O for components comprising a protein-protein complex are almost identical (grey and black linear relations). Consequently, the low resolution structure restored from a SANS with contrast variation experiment will reflect the shape of the whole complex (grey surface representation). B. Isotopic labelling of a component with non-exchangeable deuterium has a dramatic effect on the contrast relationships and the separation of match points for the individual components and of the whole complex. In this example, component 1 is labelled on average with 60% non-exchangeable 2H (grey) while component 2 remains as a native 1H-protein (orange). When the scattering contributions of the native 1H-protein are matched out (~40% v/v 2H2O; Δρ2 = 0; orange line), the scattering intensities will be derived from 2H-component 1, the magnitude of which will be proportionate to Δρ12 and V12 and P1(q). On increasing the % v/v 2H2O even further a point is reached when Δρ for the whole complex limits to zero (~75% v/v; black line) whereby the scattering signal will be exceptionally weak (essentially incoherent scattering and scattering from 1H-2H exchange). Eventually 2H-component 1 will be matched out at high % v/v 2H2O (~91% v/v 2H2O;Δρ1 = 0; grey line) leaving coherent scattering contributions from the 1H-component 2 (proportionate to Δρ22 and V22 and P2(q)). From a set of contrast variation data it is possible to determine the shapes of the entire complex, of the individual components and the orientations of the components within the complex.