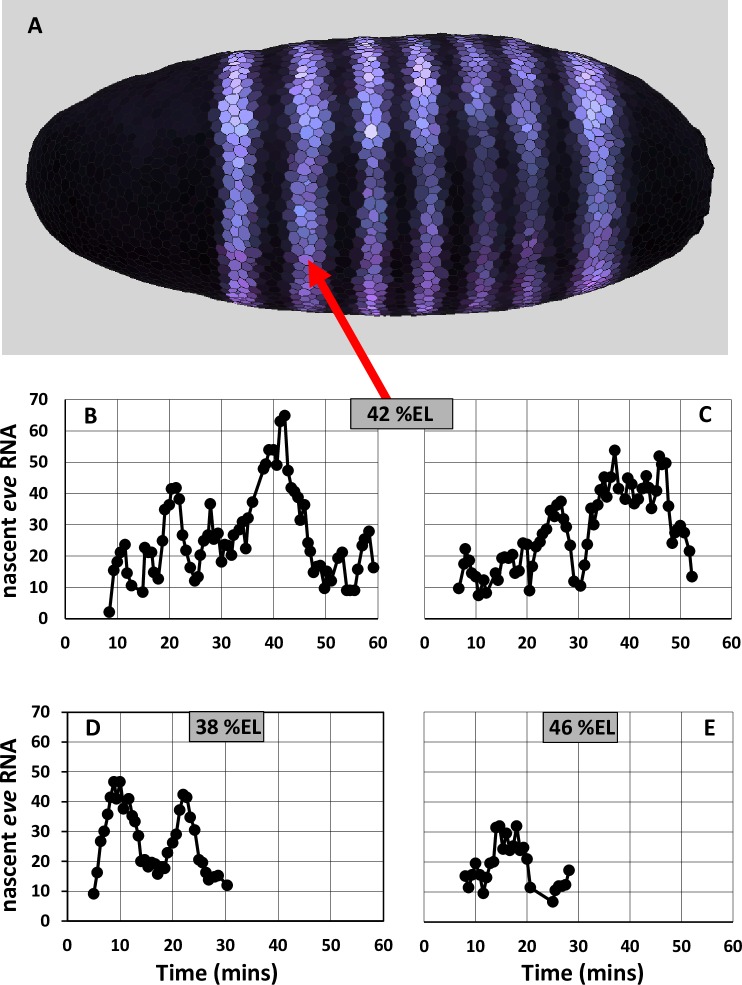
Fig 1. mRNA expression for the pair-rule gene even-skipped (eve) in Drosophila.
(A) Spatially, eve (purple) is expressed in 7 stripes orthogonal to the anterior-posterior (AP) axis, each 5–10%EL (percent embryo length) wide. Anterior is to the left. Data from the BDTNP database (http://bdtnp.lbl.gov/Fly-Net/), nuclear and cytoplasmic staining for eve mRNA in embryo 11081-02jn060-06 at 40% membrane invagination (mid nuclear cleavage cycle 14), projected onto a canonical embryo geometry. Image created with PointCloudXplore. Expression at stripe 2, centered at approximately 42%EL from the anterior pole (red arrow), can be driven by the minimal stripe element (MSE). (B—E) Time series of the number of nascent eve transcripts in stripe 2 show stochastic gene expression at per minute resolution. (B, C) Time series from two individual nuclei at stripe center (approx. 42%EL), data from Fig 4A and S2B Fig respectively of [4]. (D) Time series from a nucleus on the anterior edge of stripe 2 (approx. 38%EL), data from S2ig of [4]. (E) Time series from a nucleus at the posterior edge of stripe 2 (approx. 46%EL), data from S2D Fig of [4].