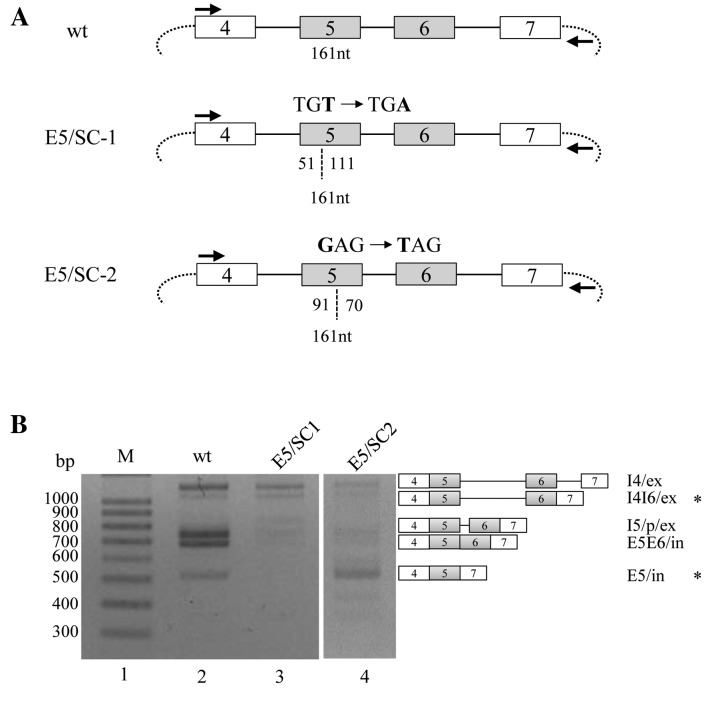
Figure 1.
Various locations of the stop-codon in exon 5 have various effects on the degradation of the spliced and partially spliced isoforms from RON pre-mRNA. (A) The construction of the E5/SC-1 (T-to-A substitution 51 nucleotides downstream from the 3′ splice-site of exon 5) and E5/SC-2 (G-to-T substitution 91 nucleotides downstream from the 3′ splice-site of exon 5) minigenes is shown. Exons are indicated with boxes; introns are marked with lines; vector sequences are presented with arcs. The locations of the stop-codons are shown with the numbers of nucleotides. Primers used for RT-PCR analysis are indicated with arrows. (B) Results of the RT-PCR analysis of E5/SC-1 and E5/SC-2 minigenes are presented. The identities of each of the isoforms are indicated at the right panel. The isoforms observed to not decrease are designated with asterisks. RON, récepteur d'origine nantais; RT-PCR, reverse transcription-polymerase chain reaction; wt, wild type; M, molecular weight ladder; I4/ex, intron 4 is spliced; I4I6/ex, introns 4 and 6 are spliced; I5/p/ex, cryptic splice site in intron 5 is activated; E5E6/in, exons 5 and 6 are included; E5/in, exon 5 included and exon 6 excluded.