Abstract
The potential prognostic significance of DNA flow cytometric measurements (DNA ploidy and S-phase fraction) in breast cancer remains in dispute. Inconclusive data, primarily due to the lack of consistent standardization and quality control programs, have limited its translation into clinical practice. The aim of the present review, based on the 25-year experience of the Portuguese Institute of Oncology of Lisbon, is to assess the clinical relevance and application of DNA flow cytometry for the prognosis of breast cancer. Overall, data from Portuguese Institute of Oncology of Lisbon indicate that DNA flow cytometry provides significant prognostic information that is biologically relevant and clinically useful for the management of patients with breast cancer. Furthermore, this data has demonstrated the independent value of DNA aneuploidy as a prognostic indicator of poor clinical outcome in various subgroups of patients with early or locally advanced breast cancer at short- and long-term follow-up. Notably, aneuploidy identifies subsets of patients with grade (G)1 or G2 tumours who exhibit a poor clinical outcome. These patients may benefit from adjuvant chemotherapy, particularly those with luminal A and luminal B/human epidermal growth factor-2-negative endocrine-responsive breast cancer. In conclusion, data from Portuguese Institute of Oncology of Lisbon reinforces the clinical importance and utility of DNA flow cytometric analysis, particularly DNA ploidy, in the prognostic assessment and therapeutic planning for patients with breast cancer.
Keywords: breast cancer, DNA flow cytometry, DNA ploidy, S-phase fraction, prognosis
1. Introduction
Invasive breast carcinoma is one of the most frequent types of malignant neoplasm and the leading cause of cancer-associated mortality in females (1). The clinicopathological characteristics typically used to assess prognosis and guide therapy for this disease frequently fail to characterize the multiple aspects and clinical heterogeneity of breast cancer, particularly concerning the biological behaviour of tumours in individual cases.
DNA flow cytometry is a widely-used technique to estimate the DNA content (DNA ploidy) and proliferative activity (S-phase fraction; SPF) in a number of human neoplasms. In breast cancer, DNA ploidy and SPF have been investigated in association with the biological characterization of tumours, patient survival and prognosis (2–15).
Numerous studies have reported that DNA ploidy and/or SPF are prognostic markers for breast cancer (2–11); however, a number of studies have questioned their independent prognostic significance (12–15). These conflicting data have limited the prognostic value of DNA flow cytometry and the extent of its clinical use. The primary reason for the discrepant data appears to be a lack of consistent standardization and quality control programs to ensure the intra- and inter-laboratory reproducibility of results (5,16–20). In addition, intratumoral biological heterogeneity (21,22) and SPF categorization (16,17) have been implicated in the variability of the results. However, despite the inconclusive data on the prognostic value of DNA flow cytometry, numerous hospitals and cancer centres perform cytometric analyses to aid in the measurement of prognostic markers for breast cancer (23).
Previous studies have indicated that the evaluation of cytometric parameters can improve the management of breast cancer, aiding in the prognostic assessment and therapeutic planning for patients (2–6,9,10,24). The current study aimed to review the application of DNA flow cytometry in the prognostic assessment of patients with breast cancer. The present review is based on a 25-year experience of using DNA cytometric technology at the Portuguese Institute of Oncology of Lisbon (Lisbon, Portugal).
2. Patients and methods
Patients
DNA flow cytometry has been routinely used at the Portuguese Institute of Oncology of Lisbon since 1990, to evaluate the surgical specimens and fine-needle cytological aspirates of patients with breast cancer. Patients (females; n>1000; age range, 23–91 years old) in the present review were diagnosed and treated at the Portuguese Institute of Oncology according to standardized therapeutic protocols between August 1990 and April 2007.
Therapies
Patients in the previous studies that the current report is reviewing received a modified radical mastectomy or conservative treatment with an axillary lymph node dissection. All lymph node-positive premenopausal patients, hormone receptor-negative postmenopausal patients and lymph node-negative patients with a high-risk of tumour recurrence received adjuvant chemotherapy. Patients with a tumour >2 cm, a poorly differentiated tumour or hormone receptor-negative tumour were considered to have a high-risk of tumour recurrence. Endocrine treatment was recommended for all hormone receptor-positive patients with breast cancer. Patients who underwent conservative breast surgery received local radiotherapy postoperatively, while lymph node metastasis-positive patients received radiotherapy to the axillary region. The majority of the changes to the therapeutic system in place to treat patients with breast cancer at the Portuguese Institute of Oncology of Lisbon were associated with the introduction of taxanes in 1998 and trastuzumab in 2005. No patients had metastatic breast cancer at time of diagnosis or had received treatment prior to surgical resection. All studies reviewed in the present report were approved by the Portuguese Institute of Oncology Institutional Review Board and Ethics Committee.
DNA flow cytometry
Based on the experience at the Portuguese Institute of Oncology of Lisbon, the key to performing accurate and reproducible DNA flow cytometry assessments is based on the following methodological factors: i) The preferential use of fresh/frozen tumour samples, since on formalin-fixed paraffin-embedded tissue, particularly for SPF determination, the DNA histograms demonstrate low technical quality and problems with standardization; ii) the examination for the presence of tumour cells in the tissue sample using haematoxylin and eosin staining of a cell suspension aliquot obtained following mechanical/enzymatic disaggregation; iii) the adequate internal standardization with chicken/trout red blood cells to aid in the localization of the diploid reference peak, primarily consisting of fibroblasts, lymphocytes and healthy epithelial cells in the sample; iv) the optical alignment of the cytometer with calibrated fluorospheres; v) the acquisition of ≥15,000 nuclei at a slow rate; vi) the exclusion of DNA histograms with coefficients of variation for tumour G0/G1 peaks >6% (5,16–20).
Regarding DNA ploidy, tumours were categorized according to the DNA index (DI) as follows: Hypodiploid, DI<0.95; diploid, DI=0.95–1.05; hyperdiploid, DI>1.05–1.92; tetraploid, DI>1.92–2.04; hypertetraploid, DI>2.04; and multiploid, ≥2 DIs (7). SPF is a continuous biological variable rather than a dichotomous function; thus, each laboratory should establish its own SPF data (18). The current study used a median SPF value of 6.1% as the cut-off point to distinguish between tumours with low and high proliferative activity, as when distinct SPF variables were previously investigated, this cut-off value provided the highest prognostic significance (25). As recommended by previous studies (18–20), another approach to reporting SPF is the subdivision into tertiles (<4.5, 4.5–9.2 and ≥9.2%), which has been proven to possess prognostic significance (25). However, as reported previously, a well-known limitation of SPF is the fact that in 20–25% of cases, primarily those with aneuploidy, determining SPF from the DNA histogram fails for the following technical reasons: i) Overlap of populations; ii) a high degree of background debris; iii) too few cells; and iv) tumour classification as multiploid and/or hypertetraploid (9,10).
Survival analysis
In previous studies performed at the Portuguese Institute of Oncology of Lisbon, there was systematic focus on the application of survival analysis methods, including the following: Sample size; length of follow-up time; definition of the end points of interest; and the choice and quality of the univariate, multivariate and graphical analyses (7,8,11). The statistical power of survival studies depends on the accurate assessment of these variables, in order to prevent statistical inadequacies (26). The cumulative probability of patient survival was evaluated using the Kaplan-Meier estimator. The differences between survival curves was analysed using the log-rank test. The relative strength and independent prognostic value of the variables investigated was analysed using Cox's proportional hazard regression model analysis. P<0.05 was considered to indicate a statistically significant difference. R software (version 3.3.0; R Foundation for Statistical Computing, Vienna, Austria) was used to perform statistical analyses.
The clinical data from the follow-up of patients was obtained from the review of clinical records or consultation of the epidemiological registry service at the Portuguese Institute of Oncology of Lisbon. Disease-free survival (DFS) was defined as the time elapsed between diagnosis and documented local or distant tumour recurrence. Overall survival (OS) was defined as the time interval between diagnosis and mortality from breast cancer.
3. Association between DNA ploidy/SPF, and the histopathological and molecular characteristics frequently associated with breast cancer prognosis
A consistent result from our previous cytometric studies was the significant correlation between DNA aneuploidy/high SPF and a high grade of tumour differentiation (P<0.001) (7,8,11), greater tumour size (P<0.002) (7), lack of hormone (oestrogen and progesterone) receptors (P<0.001) (7,8,11) and human epidermal growth factor expression-2 (HER-2; P<0.001) (27). In addition, no significant association between DNA ploidy/SPF and axillary lymph node status was identified (7,8,11,27). Relevant findings are summarized in Table I.
Table I.
Data on the application of DNA flow cytometric measurements as prognostic markers for patients with breast cancer at the Portuguese Institute of Oncology of Lisbon.
| Sample type | No. of patients | Median follow-up (months) | Data presented | Reference studies |
|---|---|---|---|---|
| Fresh/frozen | 308, 392, 393 and 306, respectively | 39.6, 81, 134 and 39.6, respectively | Aneuploidy and high SPF are significantly associated with high tumour grade, a greater tumour size, lack of hormone receptors and HER-2 expression | (7,8,11,25) |
| Fresh/frozen | 308 | 39.6 | At short term follow-up, axillary nodal status, DNA ploidy (for OS) and SPF (for DFS) demonstrated independent prognostic significance through multivariate analysis | (7) |
| Fresh/frozen | 308 | 39.6 | At short term follow-up, aneuploidy and high SPF were significantly associated with poor prognosis in axillary node-positive disease | (7) |
| Fresh/frozen | 308, 393 and 189, respectively | 39.6, 134 and N/A, respectively | Multiploid/hypertetraploid tumours demonstrated the poorest clinical evolution | (7,11,30) |
| Fresh/frozen | 306 | 39.6 | SPF is a better prognostic indicator for breast cancer compared with Ki-67 expression | (25) |
| Fine-needle cytological aspirates | 392 | 81 | In advanced breast cancer, disease stage, absence of progesterone receptors and DNA aneuploidy are independently associated with poor OS | (8) |
| Fresh/frozen | 393 | 134 | At long term follow-up, aneuploidy is an indicator of poor prognosis in patients with axillary node-positive disease and G2 tumours, and an indicator for poor DFS in axillary node-negative patients | (11) |
| Fresh/frozen | 684 | 134.5 | Aneuploidy identifies subsets of patients with early breast cancer (G1 and G2 tumours) who demonstrated poor clinical outcome | (32) |
SPF, S-phase fraction; HER-2, human epidermal growth factor 2; OS, overall survival; DFS, disease-free survival; G, grade; N/A, not applicable.
4. Independent prognostic value of DNA ploidy and SPF in breast cancer compared with established prognostic factors
In a study of 308 patients with breast cancer (7), with a short-term follow-up (median follow-up time, 39.6 months), Cox's proportional hazard regression model analysis was used to investigate the disease staging (tumour-node-metastasis) (28), histopathological tumour grading (Nottingham Histologic Score system) (29), DNA flow cytometry results, oestrogen and progesterone receptor expression status, and marker of proliferation Ki-67 (Ki-67) and HER-2 immunohistochemical expression status. Univariate analysis (Cox's proportional hazard regression model analysis) revealed that there was a significant difference between all the above mentioned prognostic factors, excluding Ki-67 expression status (7). Multivariate analysis (Wald test) demonstrated that axillary lymph node status (OS, P=0.001; DFS, P<0.001), DNA ploidy [OS relative risk (RR), 3.43; P=0.006], and SPF (DFS RR, 4.53; P<0.001) retained independent prognostic significance (7).
5. Prognostic significance of cytometric parameters regarding the axillary node status of patients with breast cancer
In a subset analysis performed in a previous study (7), aneuploidy (P=0.005) and a high SPF (P<0.001) were significantly associated with poor DFS and OS survival in patients with axillary node-positive breast cancer. No significant differences were identified between a higher frequency of aneuploidy/a high SPF and a shorter survival time in patients with axillary node-negative breast cancer (7).
6. Prognostic significance of specific DNA aneuploidy subcategories in breast cancer
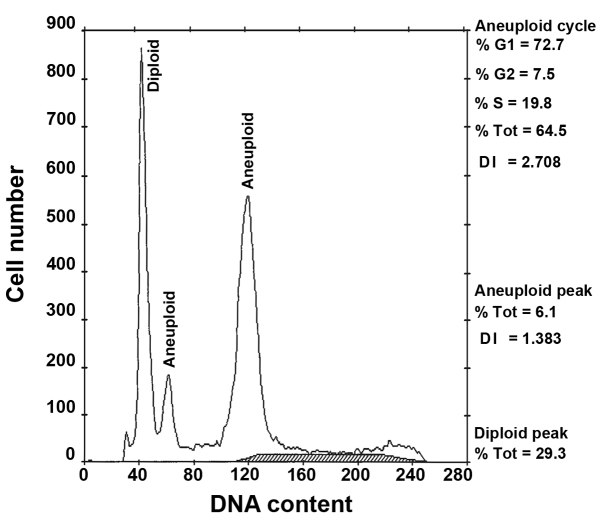
In several of our previous studies (7,11,30), patients with aneuploidy tumours, including hyperdiploid/tetraploid tumours (OS RR, 1.96; P=0.003; DFS RR, 1.70; P=0.006) and multiploid/hypertetraploid tumours (OS RR, 2.91; P<0.001; DFS RR, 2.31; P=0.003) demonstrated a significantly poorer prognosis compared with patients with DNA diploid tumours (11). In addition, patients with multiploid/hypertetraploid tumours (Fig. 1) had an increase in tumour biological aggressiveness and a poor clinical evolution compared with patients with diploid and hyperdiploid/tetraploid tumours. To investigate the underlying genetic mechanisms associated with the development of this aneuploidy, copy number variations in a cohort of patients with multiploid/hypertetraploid tumours (n=13) were analysed using comparative genomic hybridization (31). A high incidence of genetic aberrations, primarily associated with chromosomal gains was identified in this group and the 7q locus appeared to be a potential site for a number of candidate oncogenes (31).
Figure 1.
Representative DNA histogram of a multiploid tumour with an aneuploid population (DNA index 1.38) and hypertetraploid clone (DNA index 2.71). Tot, total; DI, DNA index.
7. Prognostic value of SPF compared with Ki-67 expression in breast cancer
In a study of 306 patients with primary operable breast cancer (disease stages I and II), flow cytometry and immunohistochemistry analysis were simultaneously performed in 230 cases (25). Cut-off points of 6.1% for SPF and 10% for Ki-67 expression were used to discriminate between tumours with low and high proliferation. An agreement in the proliferation data between the two methods was identified in 69.1% of cases (K statistic=0.384). However, as a cell proliferation variable SPF, but not Ki-67 expression, demonstrated prognostic significance in regards to OS (P<0.0003) and DFS (P<0.0001) (25). Therefore, SPF appears to be a better prognostic indicator for breast cancer compared with Ki-67 expression (25).
8. Prognostic value of DNA flow cytometry in locally advanced breast cancer using fine-needle aspiration cytology (FNAC)
In a prospective study (8) involving 392 patients with advanced breast cancer (stage IIB, n=106; stage IIIA, n=66; stage IIIB, n=174; stage IV, n=46) and a median follow-up time of 81 months, multivariate analysis (Cox's proportional hazard regression model analysis; Wald test) of OS using FNAC revealed that an advanced disease stage (P<0.001), the absence of progesterone receptors (P=0.008) and DNA aneuploidy (P=0.016) had independent prognostic significance (8).
9. Long-term prognostic significance of cytometric parameters in breast cancer
Due to previous results regarding the prognostic value of DNA flow cytometry in patients with breast cancer with a short-term follow-up (7), a study of 393 patients with invasive ductal breast carcinoma with a long-term follow-up (median follow-up time, 134 months) (11) was performed to evaluate ‘classical’ and cytometric prognostic parameters. In multivariate analysis (Cox's proportional hazard regression model analysis; Wald test), axillary node involvement (OS and DFS, P<0.001), DNA aneuploidy (OS, P=0.002; DFS, P=0.001) and lack of progesterone receptor expression (OS, P=0.029) demonstrated statistically significant associations with a shorter patient survival time. In axillary node-negative patients, DNA aneuploidy (DFS, P=0.030) and absence of oestrogen receptor expression (OS, P=0.010) significantly predicted poor survival. In axillary node-positive patients (OS, P=0.010; DFS, P=0.051) and those with moderately differentiated tumours [grade (G)2; OS, P=0.045; DFS, P=0.011] aneuploidy was an indicator of poorer prognosis.
10. Identification of different prognostic subgroups in patients who present with similar grades of tumour differentiation using DNA ploidy
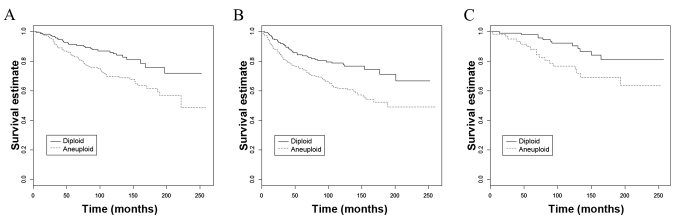
The results of a previous study (11) led to an investigation into the association between the grade of tumour differentiation and DNA ploidy in a large cohort of 684 patients with invasive breast carcinoma with a long-term follow-up (32). Patients with aneuploid G2 tumours demonstrated significantly poorer DFS (P=0.001) and OS (P<0.001), and patients with aneuploid G1 tumours demonstrated significantly poorer OS (P=0.013) (Fig. 2), compared with patients with G2 and G1 diploid tumours, respectively. The same significant prognostic associations were identified in a subset of patients (n=451) with stage I/IIA breast cancer (aneuploid G2 tumour DFS, P=0.003; OS, P=0.004; aneuploid G1 tumour OS, P=0.049). These results suggest that aneuploidy identifies subsets of patients with well (G1) and moderately (G2) differentiated tumours with a poor clinical outcome.
Figure 2.
Kaplan-Meier survival curves based on DNA ploidy for patients with G2 tumours plotted for (A) overall survival (P<0.001), (B) disease-free survival (P=0.001), and for patients with G1 tumours for (C) overall survival (P=0.013).
11. Conclusions
Numerous prognosis-associated studies using DNA flow cytometry have been performed in breast cancer (2–15); however, the results are notably variable. Contradictory findings may result from differences in study design and technical, analytical and interpretative methods (15,17–20). Despite these limitations, the prognostic significance of DNA ploidy and SPF in breast cancer is relatively consensual, at least in univariate models (3,10,33). However, an important question remains in relation to the clinical utility and predictive value of DNA ploidy and SPF in the selection of adjuvant therapies.
Similarly to studies by other institutes (34,35), data from the Portuguese Institute of Oncology of Lisbon indicates that when DNA flow cytometric analysis is performed on fresh/frozen tumour samples from a large cohort, DNA aneuploidy and/or high SPF are significant poor prognostic factors for breast cancer, as determined by univariate and multivariate analyses. Michels et al (35) and Wenger and Clark (36), in their comprehensive reviews of studies involving >100 patients and using fresh or frozen material, demonstrated that 21/24 studies, with 17 using multivariate analysis, reported an association between high SPF and poor patient outcome. Regarding DNA aneuploidy, 17/23 studies, with 8 of them using multivariate analysis, identified a correlation with poor patient prognosis. However, a number of studies using state-of-the-art DNA flow cytometry, in regards to sampling size, recommended protocols and data analysis, have reported that SPF and DNA ploidy do not provide significant prognostic information in breast cancer (13,15).
Data from the Portuguese Institute of Oncology of Lisbon demonstrates that DNA aneuploidy and a high SPF, markers of genetic instability and cell proliferation, respectively, are significantly associated with established molecular features of poor patient prognosis, including a high histological grade, and the absence of hormone receptor and HER-2 expression (20,27,35). Nevertheless, the significant association of DNA aneuploidy and a high SPF with tumour size, but lack of association with lymph node status suggests that they are indicators of tumour growth rather than metastatic potential (15,37).
In our previous studies (7,11), DNA ploidy had a significant prognostic influence in short and long follow-up periods, similar to the results of another study (13). This prognostic influence was most evident in axillary node-positive patients, a subgroup with a poor prognosis. However, in the long-term follow-up study (11), DNA ploidy retained an independent prognostic value for DFS in the subset of patients with axillary node-negative breast cancer, a subset for whom it is difficult to identify significant prognostic indicators for. This finding has been corroborated by others (33,34). Therefore, it is important that DNA ploidy can aid in the prediction of recurrence risk of patients with axillary node-negative breast cancer.
Similarly to the early stages of breast cancer, the prognostic value of DNA ploidy in advanced breast cancer, using FNAC, was identified to be significant (8). Patients with diploid tumours, compared with those with aneuploid tumours, exhibited a more positive OS (8). Notably, despite the screening mammography programs implemented, there are a non-negligible number of cases of locally advanced and metastatic breast cancer, diagnosed ab initio, that still constitute a number of oncological mortality cases. FNAC has the advantage of overcoming a number of sampling disaggregation problems associated with DNA flow cytometry (38).
Notably, the subgroup of patients with relatively uncommon multiploid/hypertetraploid tumours, comprising 5–10% of all cases, had the poorest clinical outcome (7,11,30). Other studies that investigated aneuploid subsets of patients with breast cancer have demonstrated similar results (3,39,40). It is reasonable to hypothesise that the occurrence of multiple aneuploid clones and/or hypertetraploidy supports the development of a more aggressive phenotype (41). Supporting this, Duesberg et al (42) reported that the degree of genomic instability is proportional to the degree of aneuploidy, potentially leading to tumour metastasis.
The association between SPF and DNA ploidy, particularly diploidy with low SPF and aneuploidy with high SPF, is well known. Therefore, the mutual inclusion of DNA ploidy and SPF in multivariate Cox's proportional hazard regression model analysis can produce confounding results. The association between SPF and DNA ploidy can explain why, in our previous short-term follow-up study (7), DNA ploidy and SPF demonstrated independent, prognostic significance for OS and DFS, respectively. A number of studies have recommended that an adequate SPF adjustment should be made in order to eliminate the redundant association with DNA ploidy and improve the prognostic model (43). Despite their similar prognostic significance, there are three primary reasons that make DNA ploidy a more suitable cytometric parameter in routine practice: i) The possibility of assessing ploidy in formalin-fixed paraffin-embedded sample; ii) the technical inability to determine SPF in ~1/4 of cases; and iii) difficulties in the reproducibility of SPF measurements.
Even with inherent technical limitations, the importance of SPF as a prognostic cell proliferation variable cannot be underestimated. Numerous studies have reported that SPF is the most important predictor of survival in breast cancer (2,9,10,16,17). In the studies performed at the Portuguese Institute of Oncology of Lisbon, besides the prognostic significance of SPF in the subgroup of axillary node-positive patients (7), a comparison between SPF and Ki-67 expression for assessing tumour proliferative activity (25) revealed that only SPF had statistical significance for predicting disease outcome. Ki-67 expression analysis has the technical advantage of allowing the morphological evaluation of proliferating cell populations; however, there is still an important unresolved issue concerning the identification of the optimal Ki-67 cut-off value for discriminating between tumours with low and high proliferation (44,45).
Similarly to numerous studies (10,19,33,34,37,46), results from the Portuguese Institute of Oncology of Lisbon have consistently identified a strong association between the grade of tumour differentiation and DNA ploidy in breast cancer (11,32). This result is explained by the similar biological significance of these characteristics, reflecting the extent, type and complexity of genomic abnormalities of the tumour (38,47). In addition, the majority of well-differentiated G1 tumours are diploid and the majority of poorly differentiated G3 tumours are aneuploid (11,32,46). This supports the usefulness of DNA ploidy analysis as a complementary or alternative tool to conventional histological tumour grading, providing additional and less subjective prognostic information. Furthermore, the finding that aneuploidy can identify subgroups of patients with G1 and G2 tumours that have poor prognosis increases the significance of the clinical application of DNA ploidy determination (32). The clinical relevance of this finding is associated with the potential rationally-based influence of DNA ploidy on the therapeutic decisions for patients with aneuploid low/intermediate grade tumours, particularly those whose tumours are luminal A or luminal B/HER-2-negative endocrine-responsive breast cancer, who may benefit from adjuvant chemotherapies.
In conclusion, despite the growing clinical interest and potential implications for therapeutics, the majority of complex high-throughput molecular/genomic technologies (48) are not yet fully standardized or adapted to routine practice (49,50). Notably, relevant prognostic information for breast cancer could easily be obtained by relatively simple and fast DNA flow cytometry, even in cost:benefit financial terms. Investigations into the prognostic significance of DNA flow cytometric analysis for different molecular subtypes of breast cancer, including luminal A, luminal B, HER-2 positive and triple-negative, in the future will prove interesting. Although the studies performed at the Portuguese Institute of Oncology of Lisbon are robust and reliable, they are retrospective in nature. Therefore, further well-designed randomized clinical trials with a larger cohort of patients are required to confirm the accuracy and reliability of the findings discussed in the present review.
Acknowledgements
The authors would like to thank the staff at the Departments of Surgery and Pathology at the Portuguese Institute of Oncology of Lisbon for supplying and histologically analysing the tumour samples in the previous studies herein reviewed. The present study was supported in part by a grant for the ‘Prognostic and therapeutic implications of DNA flow cytometry analysis on molecular subtypes of breast cancer evaluated by immunohistochemistry’ from the Portuguese Institute of Oncology of Lisbon.
References
- 1.Ferlay J, Steliarova-Foucher E, Lortet-Tieulent J, Rosso S, Coebergh JW, Comber H, Forman D, Bray F. Cancer incidence and mortality patterns in Europe: Estimates for 40 countries in 2012. Eur J Cancer. 2013;49:1374–1403. doi: 10.1016/j.ejca.2012.12.027. [DOI] [PubMed] [Google Scholar]
- 2.Clark GM, Dressler LG, Owens MA, Pounds G, Oldaker T, McGuire WL. Prediction of relapse or survival in patients with node-negative breast cancer by DNA flow cytometry. N Engl J Med. 1989;320:627–633. doi: 10.1056/NEJM198903093201003. [DOI] [PubMed] [Google Scholar]
- 3.Camplejohn RS, Ash CM, Gillett CE, Raikundalia B, Barnes DM, Gregory WM, Richards MA, Millis RR. The prognostic significance of DNA flow cytometry in breast cancer: Results from 881 patients treated in a single centre. Br J Cancer. 1995;71:140–145. doi: 10.1038/bjc.1995.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shiao YH, Chen VW, Lehmann HP, Wu XC, Correa P. Patterns of DNA ploidy and S-phase fraction associated with breast cancer survival in blacks and whites. Clin Cancer Res. 1997;3:587–592. [PubMed] [Google Scholar]
- 5.Bagwell CB, Clark GM, Spyratos F, Chassevent A, Bendahl PO, Stål O, Killander D, Jourdan ML, Romain S, Hunsberger B, Baldetorp B. Optimizing flow cytometric DNA ploidy and S-phase fraction as independent prognostic markers for node-negative breast cancer specimens. Cytometry. 2001;46:121–135. doi: 10.1002/cyto.1097. [DOI] [PubMed] [Google Scholar]
- 6.Tsutsui S, Ohno S, Murakami S, Hachitanda Y, Oda S. Prognostic value of DNA ploidy in 653 Japanese women with node-negative breast cancer. Int J Clin Oncol. 2001;6:177–182. doi: 10.1007/PL00012103. [DOI] [PubMed] [Google Scholar]
- 7.Pinto AE, André S, Soares J. Short term significance of DNA ploidy and cell proliferation in breast carcinoma: A multivariate analysis of prognostic markers in a series of 308 patients. J Clin Pathol. 1999;52:604–611. doi: 10.1136/jcp.52.8.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pinto AE, André S, Mendonça E, Silva G, Soares J. Overall survival in advanced breast cancer: Relevance of progesterone receptor expression and DNA ploidy in fine-needle aspirates of 392 patients. Int J Biol Markers. 2003;18:7–12. doi: 10.5301/JBM.2008.512. [DOI] [PubMed] [Google Scholar]
- 9.Vielh P, Carton M, Padoy E, de Rycke Y, Klijanienko J, El-Naggar AK, Asselain B. S-phase fraction as an independent prognostic factor of long-term overall survival in patients with early-stage or locally advanced invasive breast carcinoma. Cancer. 2005;105:476–482. doi: 10.1002/cncr.21348. [DOI] [PubMed] [Google Scholar]
- 10.Gazic B, Pizem J, Bracko M, Cufer T, Borstnar S, Pohar-Marinsek Z, Us-Krasovec M. S-phase fraction determined on fine-needle aspirates is an independent prognostic factor in breast cancer-a multivariate study of 770 patients. Cytopathology. 2008;19:294–302. doi: 10.1111/j.1365-2303.2007.00528.x. [DOI] [PubMed] [Google Scholar]
- 11.Pinto AE, Pereira T, Santos M, Branco M, Dias A, Silva GL, Ferreira MC, André S. DNA ploidy is an independent predictor of survival in breast invasive ductal carcinoma: A long-term multivariate analysis of 393 patients. Ann Surg Oncol. 2013;20:1530–1537. doi: 10.1245/s10434-012-2804-6. [DOI] [PubMed] [Google Scholar]
- 12.Bergers E, van Diest PJ, Baak JP. Cell cycle analysis of 932 flow cytometric DNA histograms of fresh frozen breast carcinoma material. Correlations between flow cytometric, clinical, and pathologic variables. MMMCP Collaborative Group. Multicenter Morphometric Mammary Carcinoma Project Collaborative Group. Cancer. 1996;77:2258–2266. doi: 10.1002/(SICI)1097-0142(19960601)77:11<2258::AID-CNCR12>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 13.Bergers E, Baak JP, van Diest PJ, Willig AJ, Los J, Peterse JL, Ruitenberg HM, Schapers RF, Somsen JG, van Beek MW, et al. Prognostic value of DNA ploidy using flow cytometry in 1301 breast cancer patients: Results of the prospective multicenter morphometric mammary carcinoma project. Mod Pathol. 1997;10:762–768. [PubMed] [Google Scholar]
- 14.Chassevent A, Jourdan ML, Romain S, Descotes F, Colonna M, Martin PM, Bolla M, Spyratos F. S-phase fraction and DNA ploidy in 633 T1T2 breast cancers: A standardized flow cytometric study. Clin Cancer Res. 2001;7:909–917. [PubMed] [Google Scholar]
- 15.Prasad AR, Divine G, Zarbo RJ. Two-color, cytokeratin-labeled DNA flow cytometric analysis of 332 breast cancers: Lack of prognostic value with 12-year follow-up. Arch Pathol Lab Med. 2001;125:364–474. doi: 10.5858/2001-125-0364-TCCLDF. [DOI] [PubMed] [Google Scholar]
- 16.Bergers E, Montironi R, van Diest PJ, Prete E, Baak JP. Interlaboratory reproducibility of semiautomated cell cycle analysis of flow cytometric DNA-histograms obtained from fresh material of 1,295 breast cancer cases. Hum Pathol. 1996;27:553–560. doi: 10.1016/S0046-8177(96)90161-6. [DOI] [PubMed] [Google Scholar]
- 17.Baldetorp B, Bendhal PO, Fernö M, Alanen K, Delle U, Falkmer U, Hansson-Aggesjö B, Höckenström T, Lindgren A, Mossberg L, et al. Reproducibility in DNA flow cytometric analysis of breast cancer: Comparison of 12 laboratories' results for 67 sample homogenates. Cytometry. 1995;22:115–127. doi: 10.1002/cyto.990220207. [DOI] [PubMed] [Google Scholar]
- 18.Hedley DW, Clark GM, Cornelisse CJ, Killander D, Kute T, Merkel D. Consensus review of the clinical utility of DNA cytometry in carcinoma of the breast. Report of the DNA cytometry consensus conference. Cytometry. 1993;14:482–485. doi: 10.1002/cyto.990140505. [DOI] [PubMed] [Google Scholar]
- 19.Ormerod MG, Tribukait B, Giaretti W. Consensus report of the task force on standardization of DNA flow cytometry in clinical pathology. DNA flow cytometry task force of the European society for analytical cellular pathology. Anal Cell Pathol. 1998;17:103–110. doi: 10.1155/1998/842306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chassevent A, Jourdan ML, Ferrero-Poüs M, Colonna M, Romain S, Spyratos F, Roth H, Bolla M. Standardization and quality control in the evaluation of proliferation parameters in T1T2, N0N1, M0 breast cancer: Multicentric retrospective study II. Bull Cancer. 1999;86:685–691. (In French) [PubMed] [Google Scholar]
- 21.Kallioniemi OP. Comparison of fresh and paraffin-embedded tissue as starting material for DNA flow cytometry and evaluation of intratumor heterogeneity. Cytometry. 1988;9:164–169. doi: 10.1002/cyto.990090211. [DOI] [PubMed] [Google Scholar]
- 22.Bergers E, van Diest PJ, Baak JP. Tumour heterogeneity of DNA cell cycle variables in breast cancer measured by flow cytometry. J Clin Pathol. 1996;49:931–937. doi: 10.1136/jcp.49.11.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ross JS, Linette JP, Stec J, Ross MS, Anwar S, Boguniewicz A. DNA ploidy and cell cycle analysis in breast cancer. Am J Clin Pathol. 2003;120(Suppl):S72–S84. doi: 10.1309/QD096UGF70T5H46G. [DOI] [PubMed] [Google Scholar]
- 24.Danielsen HE, Pradhan M, Novelli M. Revisiting tumor aneuploidy-the place of ploidy assessment in the molecular era. Nat Rev Clin Oncol. 2016;13:291–304. doi: 10.1038/nrclinonc.2015.208. [DOI] [PubMed] [Google Scholar]
- 25.Pinto AE, André S, Pereira T, Nóbrega S, Soares J. Prognostic comparative study of S-phase fraction and Ki-67 index in breast carcinoma. J Clin Pathol. 2001;54:543–549. doi: 10.1136/jcp.54.7.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altman DG, De Stavola BL, Love SB, Stepniewska KA. Review of survival analyses published in cancer journals. Br J Cancer. 1995;72:511–518. doi: 10.1038/bjc.1995.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pinto AE, André S, Pereira T, Nóbrega S, Soares J. c-erbB-2 oncoprotein overexpression identifies a subgroup of estrogen receptor positive (ER+) breast cancer patients with poor prognosis. Ann Oncol. 2001;12:525–533. doi: 10.1023/A:1011163211802. [DOI] [PubMed] [Google Scholar]
- 28.Fleming ID, Cooper JS, Henson DE, Hutter RVP, Kennedy BJ, Murphy GP, O'Sullivan B, Sobin LH, Yarbro JW. AJCC Cancer Staging Manual. 5th edition. Lippincott-Raven; Philadelphia, PA: 1997. [Google Scholar]
- 29.Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: Experience from a large study with long-term follow-up. Histopathology. 1991;19:403–410. doi: 10.1111/j.1365-2559.1991.tb00229.x. [DOI] [PubMed] [Google Scholar]
- 30.Pinto AE, André S, Nogueira M, Mendonça E, Soares J. Flow cytometric DNA hypertetraploidy is associated with unfavourable prognostic features in breast cancer. J Clin Pathol. 1997;50:591–595. doi: 10.1136/jcp.50.7.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pinto AE, Roque L, Rodrigues R, André S, Soares J. Frequent 7q gains in flow cytometric multiploid/hypertetraploid breast carcinomas: A study of chromosome imbalances by comparative genomic hybridization. J Clin Pathol. 2006;59:367–372. doi: 10.1136/jcp.2005.027722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pinto AE, Pereira T, Silva GL, André S. Aneuploidy identifies subsets of patients with poor clinical outcome in grade 1 and grade 2 breast cancer. Breast. 2015;24:449–455. doi: 10.1016/j.breast.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 33.Kute TE, Russell GB, Zbieranski N, Long R, Johnston S, Williams H, Stackhouse C, Wilkins L, Evans I, Berry P, et al. Prognostic markers in node-negative breast cancer: A prospective study. Cytom Part B Clin Cytom. 2004;59:24–31. doi: 10.1002/cyto.b.20003. [DOI] [PubMed] [Google Scholar]
- 34.Duigou F, Herlin P, Marnay J, Michels JJ. Variation of flow cytometric DNA measurement in 1,485 primary breast carcinomas according to guidelines for DNA histogram interpretation. Cytometry. 2000;42:35–42. doi: 10.1002/(SICI)1097-0320(20000215)42:1<35::AID-CYTO6>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 35.Michels JJ, Duigou F, Marnay J. Flow cytometry in primary breast carcinomas. Prognostic impact of proliferative activity. Breast Cancer Res Treat. 2000;62:117–126. doi: 10.1023/A:1006451908322. [DOI] [PubMed] [Google Scholar]
- 36.Wenger CR, Clark GM. S-phase fraction and breast cancer-a decade of experience. Breast Cancer Res Treat. 1998;51:255–265. doi: 10.1023/A:1006188512927. [DOI] [PubMed] [Google Scholar]
- 37.Batsakis JG, Sneige N, el-Naggar AK. Flow cytometric (DNA content and S-phase fraction) analysis of breast cancer. Cancer. 1993;71(Suppl 6):S2151–S2153. doi: 10.1002/1097-0142(19930315)71:6+<2151::AID-CNCR2820711604>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 38.Sneige N. Utility of cytologic specimens in the evaluation of prognostic and predictive factors of breast cancer: Current issues and future directions. Diagn Cytopathol. 2004;30:158–165. doi: 10.1002/dc.20005. [DOI] [PubMed] [Google Scholar]
- 39.Michels JJ, Duigou F, Marnay J, Denoux Y, Delozier T, Chasle J. Flow cytometry in primary breast carcinomas: Prognostic impact of multiploidy and hypoploidy. Cytometry B Clin Cytom. 2003;55:37–45. doi: 10.1002/cyto.b.10040. [DOI] [PubMed] [Google Scholar]
- 40.Beerman H, Kluin PM, Hermans J, van de Velde CJ, Cornelisse CJ. Prognostic significance of DNA-ploidy in a series of 690 primary breast cancer patients. Int J Cancer. 1990;45:34–39. doi: 10.1002/ijc.2910450108. [DOI] [PubMed] [Google Scholar]
- 41.Rew DA. Significance of aneuploidy. Br J Surg. 1994;81:1416–1422. doi: 10.1002/bjs.1800811005. [DOI] [PubMed] [Google Scholar]
- 42.Duesberg P, Rausch C, Rasnick D, Hehlmann R. Genetic instability of cancer cells is proportional to their degree of aneuploidy. Proc Natl Acad Sci USA. 1998;95:13692–13697. doi: 10.1073/pnas.95.23.13692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bagwell CB. DNA histogram analysis for node-negative breast cancer. Cytometry A. 2004;58:76–78. doi: 10.1002/cyto.a.90004. [DOI] [PubMed] [Google Scholar]
- 44.Goldhirsch A, Winer EP, Coates AS, Gelber RD, Piccart-Gebhart M, Thürlimann B, Senn HJ. Panel members: Personalizing the treatment of women with early breast cancer: Highlights of the St Gallen international expert consensus on the primary therapy of early breast cancer 2013. Ann Oncol. 2013;24:2206–2223. doi: 10.1093/annonc/mdt303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tashima R, Nishimura R, Osako T, Nishiyama Y, Okumura Y, Nakano M, Fujisue M, Toyozumi Y, Arima N. Evaluation of an optimal cut-off point for the Ki-67 index as a prognostic factor in primary breast cancer: A retrospective study. PLoS One. 2015;10:e0119565. doi: 10.1371/journal.pone.0119565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frierson HF., Jr Grade and flow cytometric analysis of ploidy for infiltrating ductal carcinomas. Hum Pathol. 1994;24:24–29. doi: 10.1016/0046-8177(93)90058-O. [DOI] [PubMed] [Google Scholar]
- 47.Roylance R, Gorman P, Harris W, Liebmann R, Barnes D, Hanby A, Sheer D. Comparative genomic hybridization of breast tumors stratified by histological grade reveals new insights into the biological progress of breast cancer. Cancer Res. 1999;59:1433–1436. [PubMed] [Google Scholar]
- 48.Sotiriou C, Pusztai L. Gene-expression signatures in breast cancer. N Engl J Med. 2009;360:790–800. doi: 10.1056/NEJMra0801289. [DOI] [PubMed] [Google Scholar]
- 49.Reis-Filho JS, Westbury C, Pierga JY. The impact of expression profiling on prognostic and predictive testing in breast cancer. J Clin Pathol. 2006;59:225–231. doi: 10.1136/jcp.2005.028324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ioannidis JP, Allison DB, Ball CA, Coulibaly I, Cui X, Culhane AC, Falchi M, Furlanello C, Game L, Jurman G, et al. Repeatability of published microarray gene expression analyses. Nat Genet. 2009;41:149–155. doi: 10.1038/ng.295. [DOI] [PubMed] [Google Scholar]