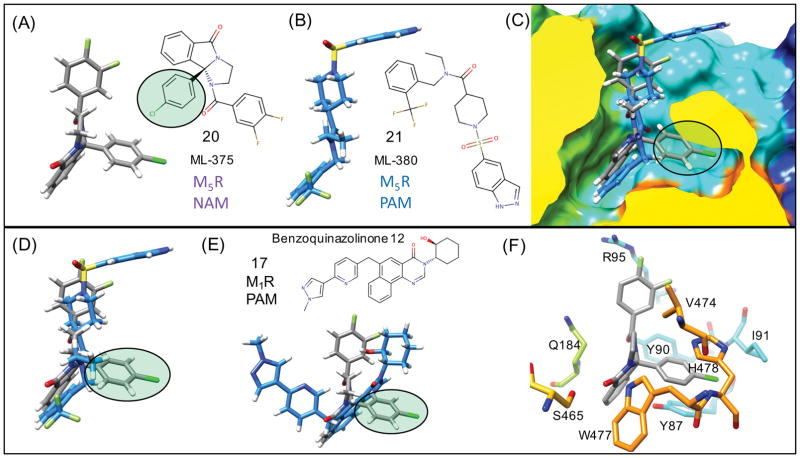
Figure 11. Flexible receptor M5R NAM unique binding mode.
Predicted lowest energy binding modes for an M5R selective NAM 20 (ML-375) (A) and PAM 21 (ML-380) (B) binding to M5R. (C) Superposition of (A) and (B) from a side view of the M5R surface shows how the p-chloro group of NAM forms unique interactions with the receptor compared to the PAM mode. (D) Only the superposition of the NAM and PAM is shown to highlight how there is minimal overlap in ligand density for the NAM (p-chloro group) with the superimposed bound PAM. (E) NAM binding mode compared with a superposition of 17 as a representative of the tricyclic M1R PAM (F) Specific residues of M5R that participate in protein-ligand interactions with the NAM, where induced-fit side chain rearrangements of Y87 and H478 are key to the interactions with the NAM (p-chloro group).