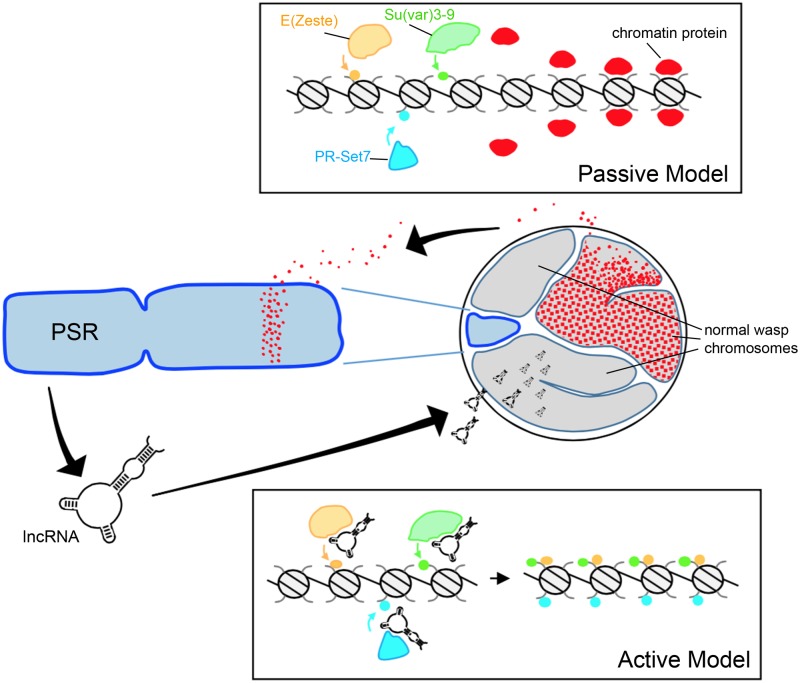
FIGURE 2.
Hypothetical models for paternal genome elimination by PSR. (Top) The passive model for genome elimination in which PSR serves as a sink, titrating away one or more chromatin-associated factors that are critical for normal chromosome condensation and resolution. Depletion of these factors allows enzymes such as E(z), Su(var)3-9, and Pr-Set7 to abnormally modify regions of chromatin within the paternal pronucleus with marks such as H3K27me1, H3K9me2,3, and H4K20me1, respectively (also see Figure 1A). In the active model (Bottom), one or more PSR expressed factors actively interferes with the chromatin remodeling process. In this example, a PSR-expressed long non-coding RNA (lncRNA) associates with the euchromatin of the wasp’s normal chromosomes (but not PSR). There, these factors inappropriately recruit chromatin-modifying enzymes to these regions, marking the normal chromosomes for elimination. Either of these models could potentially take place during spermatogenesis or in the egg cytoplasm, immediately before the first embryonic mitosis.