Figure 2.
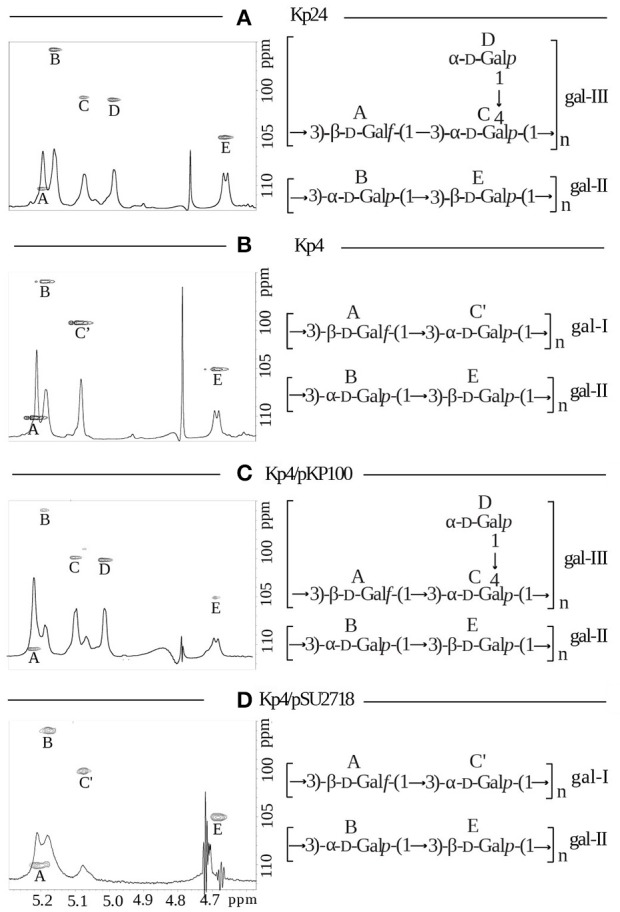
Structural analysis of lipopolysaccharides from K. pneumoniae strains: (A) Kp24 (gml+), (B) Kp4 (gml–), (C) Kp4 transformed with pKP100 (recombinant plasmid carrying gmlABC), and (D) Kp4 transformed with pSU2718 (empty cloning vector). Anomeric regions of 1H, 13C HSQC NMR spectra overlaid with 1H NMR profiles of the isolated O-PS for strains Kp24 (A), Kp4 (B), Kp4/pKP100 (C), and intact LPS for the strain Kp4/pSU2718 (D) (left panels). NMR spectra for the strain Kp4/pSU2718 were acquired using HR-MAS technique at a spin rate of 4 kHz at 30°C. The capital letters refer to carbohydrate residues as described in Tables 1, 2. For each strain O-PS gal-I and gal-II repeating units were shown as inset structures (right panels).
