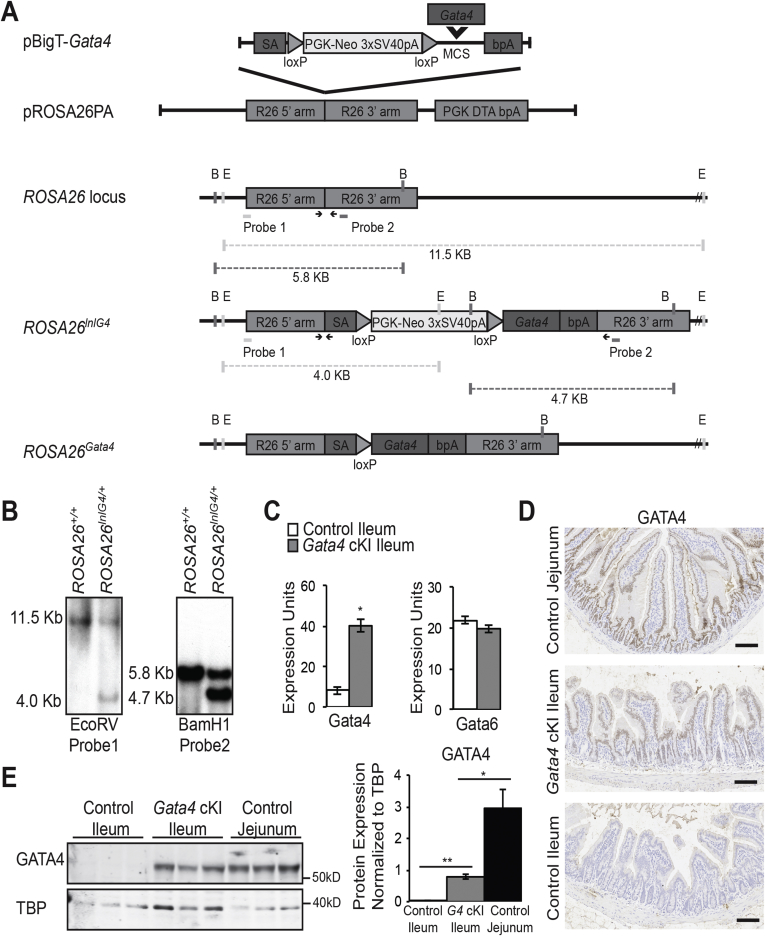
Figure 1.
Gata4 conditional knock-in mice express GATA4 in the ileum. (A) Schematic illustrating the strategy used to generate a conditional Gata4 knock-in mouse line. The coding sequence of the mouse Gata4 gene was amplified by PCR and inserted into XhoI/SacI sites in the multiple cloning site (MCS) of pBig-T to generate pBigT-Gata4. The targeting cassette consisting of an adenoviral splice acceptor (SA), a loxP flanked phosphoglycerate kinase (PGK) promoter-neomycin resistance gene (Neo) and 3×SV40 polyadenylation sequence (pA) sequence (loxP-PGK-Neo-3×SV40pA-loxP, LNL), the Gata4 coding sequence, and a bovine growth hormone polyadenylation (pA) sequence was excised from pBigT-Gata4 with PacI/AscI and inserted into the PacI/AscI sites in pROSA26PA to create pROSA26PA-Gata4. Homologous recombination between pROSA26PA-Gata4 and the endogenous ROSA26 locus in mouse R1 embryonic stem cells yielded the targeted locus Gt(ROSA)26Sortm1(Gata4)Bat, designated ROSA26lnlG4. After Cre recombination to excise the LNL cassette, Gata4 is expressed. BamHI (B) and EcoRV (E) restriction sites used for Southern blot analysis, the position of Southern blot probes, and relevant BamHI and EcoRV restriction digest fragments identified by Southern blot are shown. Arrows mark sites of genotyping primers (Table 1, primers). (B) Southern blot analysis confirmed germline transmission of the ROSA26lnlG4 allele. Representative Southern blot analysis of EcoRV or BamHI digested genomic DNA harvested from a wild-type mouse (ROSA26+/+) or a mouse heterozygous for the modified ROSA26 allele (ROSA26lnlG4/+). We observed the expected fragments representing the wild-type and modified alleles (EcoRV digest, 11.5-kb wild-type allele and 4.0-kb modified allele; BamHI digest, 5.8-kb wild-type allele and 4.7-kb modified allele). (C) qRT-PCR showed that Gata4 mRNA was induced in ileum of ROSA26lnlG4/+Villin-Cre (designated Gata4 cKI) mice compared with ileum of control mice (ROSA26lnlG4/+). Gata6 mRNA remained unchanged in the ileum of Gata4 cKI mice compared with controls (n = ileum of 5 control and 6 Gata4 cKI animals; experiments performed in triplicate). Glyceraldehyde-3-phosphate dehydrogenase was used for normalization. Error bars show SEM. P values were determined by 2-sample Student t test: *P ≤ .05. (D) Immunohistochemistry showed nuclear GATA4 protein (brown staining) in ileal epithelium of Gata4 cKI mice and in the jejunal epithelium of control mice whereas GATA4 protein was absent from ileal epithelium of control mice. Sections from at least 3 control and 3 Gata4 cKI animals were evaluated. Hematoxylin was used to counterstain tissue. Scale bars: 100 μm. (E) Immunoblot analysis of nuclear extracts from jejunal and ileal epithelial cells of control mice and from ileal epithelial cells of Gata4 cKI mice was used to quantify GATA4 protein in ileum of Gata4 cKI mice and to compare GATA4 abundance between control jejunum and GATA4-expressing ileum. The blot shown contains nuclear protein extracts from 3 control and 3 Gata4 cKI animals and is representative of analysis of more than 24 control and Gata4 cKI animals. To quantify protein expression, signal was measured using quantitative infrared immunoblotting (LI-COR) and National Institutes of Health ImageJ software. GATA4 protein levels were normalized to TATA binding protein (TBP) levels. GATA4 expression in ileum of Gata4 cKI mice was 27% the level observed in control jejunum. Molecular weight marker locations are indicated. Error bars show SEM. P values determined by 2-sample Student t test: *P ≤ .05, **P ≤ .001.