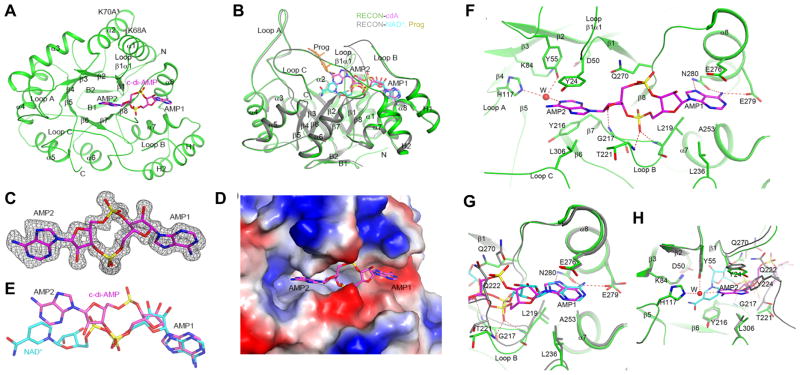
Figure 2. C-di-AMP blocks the NAD+ and substrate binding sites of RECON.
(A) Schematic drawing of structure of RECON (green) in complex with c-di-AMP (magenta, stick models). The view is down the central barrel of the structure. The two Lys to Ala mutation sites are labeled.
(B) Overlay of structure of RECON (green) in complex with c-di-AMP (magenta, stick models) and that of RECON (gray) in complex with NAD+ (cyan, stick models) (PDB entry 3LN3). The view is related to that of panel A by a 90° rotation around the horizontal axis. The bound position of the progesterone substrate (Prog) to AKR1C1 is also shown (orange, stick models) (Couture et al., 2003).
(C) Side views of (A) with two residue-mutations indicated. Omit Fo–Fc electron density for c-di-AMP at 1.5 Å resolution, contoured at 2.5σ.
(D) Molecular surface of RECON near c-di-AMP, colored by electrostatic potential.
(E) Comparison of the binding modes of c-di-AMP (magenta) and NAD+ (cyan) to RECON. All structure figures were produced with PyMOL (www.pymol.org).
(F) Detailed interactions between c-di-AMP and RECON. Hydrogen-bonding interactions are indicated with dashed lines (red). Residues involved in interactions with c-di-AMP are labeled. A water molecule is shown as a red sphere and labeled W.
(G) Overlay of the binding sites of NAD+ (cyan) and c-di-AMP (magenta) near the first nucleotide (AMP1) of c-di-AMP.
(H) Overlay of the binding sites of NAD+ (cyan) and c-di-AMP (magenta) near the second nucleotide (AMP2) of c-di-AMP.
See also Table S1.