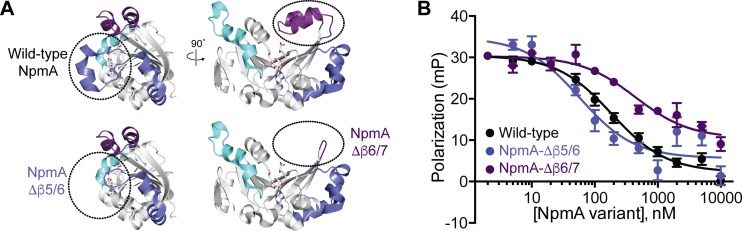
FIG 4.
Deletion of the NpmA β5/6 or β6/7 linker has opposite impact on 30S-NpmA affinity. (A) Wild-type NpmA structure shown in two orthogonal views (top) and the remaining structure after deletion of the β5/6 linker (bottom left) or β6/7 linker (bottom right) regions. Color coding of the NpmA linkers and the right-side view of wild-type NpmA are the same as those shown in Fig. 1B. (B) Competition FP binding experiments with NpmA-E184C* and unlabeled NpmA linker deletion variants, NpmA-Δβ5/6 and NpmA-Δβ6/7. The wild-type NpmA data, shown for comparison, are the same as those shown in Fig. 2. Error bars represent the SEM. Binding affinities (Ki) for each variant protein derived from these data are shown in Table 1.