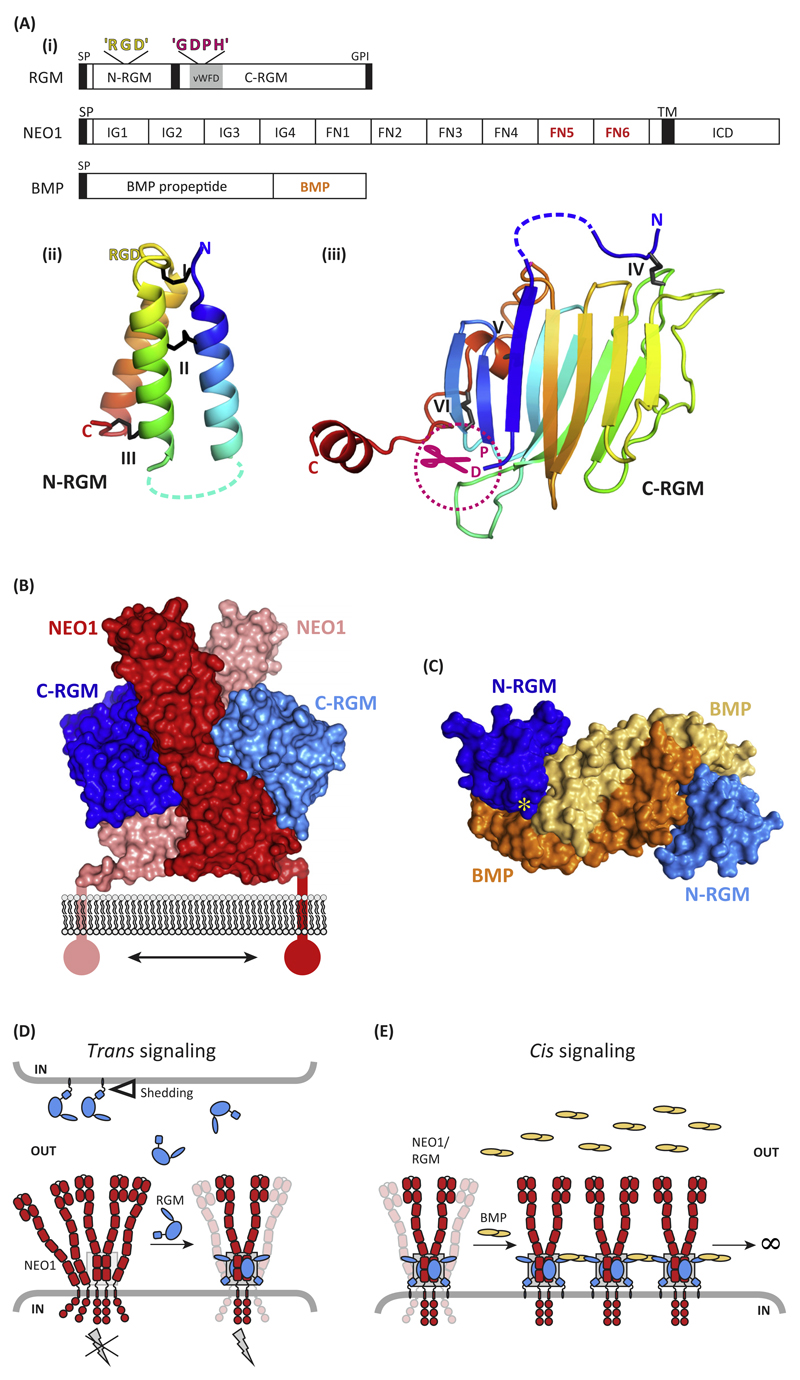
Figure 1. Molecular Determinants of Repulsive Guidance Molecules (RGMs) and Their Interactions with Neogenin (NEO1) and Bone Morphogenetic Protein (BMP) Ligands.
(A) (i) Schematic representation and domain organization of RGMs, Neogenin, and BMPs. Both the N-terminal domain (ii) [N-RGM; Protein Data Bank (PDB) ID 4UI1] and C-terminal domain (iii) (C-RGM; PDB ID 4BQ6) form distinct domains stabilized by several intramolecular disulfide bonds. Structures are shown in cartoon in rainbow coloring (blue, N terminus; red, C terminus). Note that RGD motifs are potential integrin-binding sites, but that no binding of RGMs to integrins has been reported so far. (B) The RGM–Neogenin complex (PDB ID 4BQ6). Two C-RGM molecules (blue) act as a molecular staple bringing together two Neogenin receptors (red). (C) The N-RGM–BMP complex (PDB ID 4UI1). The disulfide-linked BMP2 dimer binds two molecules of N-RGM in its wing region. The yellow asterisk indicates the position of the ‘RGD’ motif. (D) Model for RGM-mediated signaling in trans. The RGM ectodomain can be shed by proteolytic or phospholipase activity (open triangle). RGM binding to preclustered Neogenin results in stabilization and dimerization of the Neogenin ectodomain, subsequently activating downstream signaling (gray lightning bolt). The gray box highlights the RGM–Neogenin signaling hub observed in the crystal structure. (E) Model of RGM-mediated signaling in cis. RGMs can act as a physical protein bridge bringing together Neogenin and the BMP ligand, resulting in clustering. Abbreviations: FN, fibronectin; GDPH, autoproteolysis cleavage motif; GPI, glycosylphosphatidylinositol anchor; ICD, intracellular domain; IG, immunoglobulin; L, flexible linker; RGD, Arg-Gly-Asp; SP, signal peptide; TM, transmembrane; vWFD, von Willebrand Factor type D.