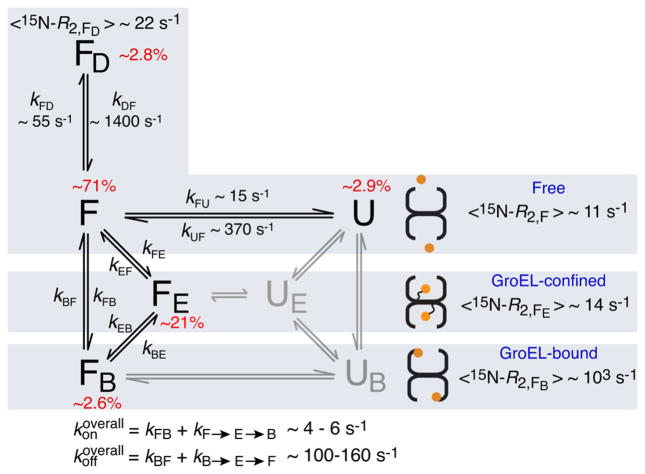
Figure 3.
Kinetic scheme for the interaction of SH3MutΔ56 with apo GroEL. The populations of states UE and UB, colored gray, are below the limits of detection and were therefore not included in the fitting of the relaxation data (see the Supporting Information for theory and details of the fitting procedure). The rate constants and populations relate to experimental conditions of 100 μM SH3 domain and 120 μM (in subunits) GroEL at 10 °C. Binding of the dimer FD to GroEL is assumed to be undetectable because of both its size and the fact that dimerization occludes the GroEL-binding surface (Figure S6). For SH3WTΔ57, there is no evidence of the existence of an unfolded state as no relaxation dispersion is observed. The 15N ΔR2 and DEST data for SH3WTΔ57 (Figure S7) show binding of the folded state to GroEL and are explained by two-state exchange between states F and FB, with kFB and kBF values of ~7 and 500 s−1, respectively (corresponding to pB ~ 1.4%).