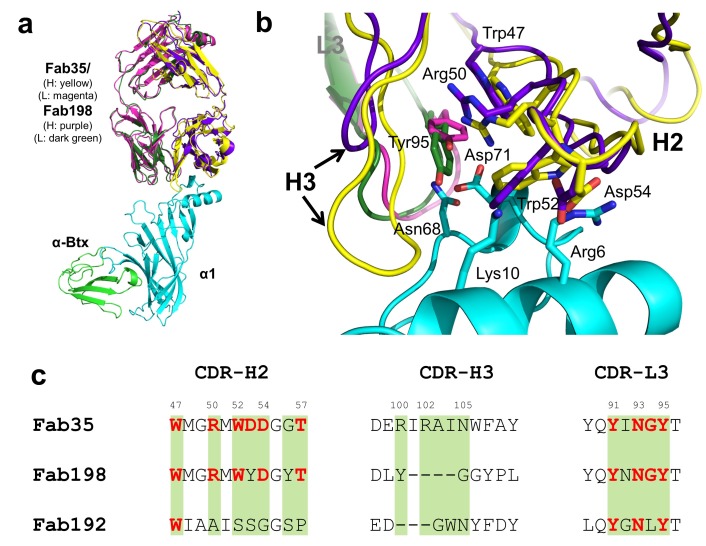
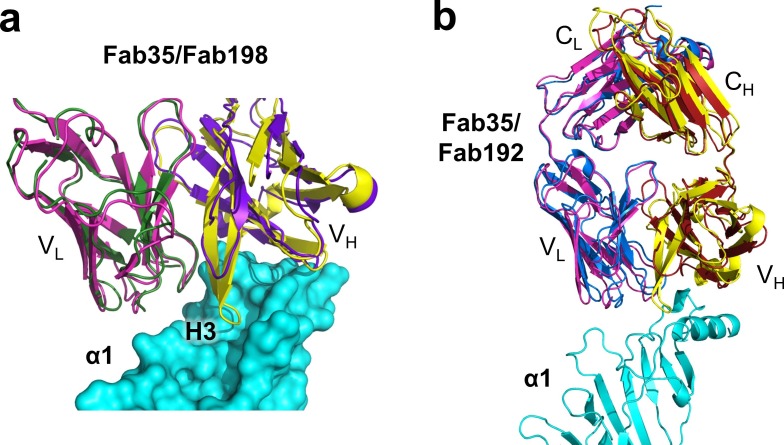
Figure 4. Structural comparisons among MG mAbs.
(a) Superposition of Fab198 (Poulas et al., 2001) (heavy chain: purple and light chain: dark green) onto Fab35 in the Fab35/nAChR α1/α-Btx ternary complex using the Cα backbone. (b) Detailed comparison of the binding interface. The residues are colored according to their protein subunits. Note that key α1-binding residues in Fab35, including Trp47, Arg50, Trp52 and Asp54 of VH and Tyr95 of VL are conserved in Fab198, and seem to be able to make similar contacts to nAChR α1 in the modeled interface. The CDR-H3 loop of Fab198 (purple) is substantially shorter than that of Fab35 (yellow), as indicated by arrows. (c) Structure-based sequence alignment of the nAChR α1-binding loops (CDR-H2, CDR-H3 and CDR-L3) between Fab35, Fab198 and Fab192 (Kontou et al., 2000). Residues shaded in light green are involved in nAChR α1 binding in Fab35, some of these (in bold font and colored in red) are conserved in Fab198 or Fab192. Note that Fab35 and Fab198 share a high similarity in their nAChR α1-binding CDR-H2 and CDR-L3 loops, but differ significantly in CDR-H3. On the other hand, Fab192 differs significantly from Fab35 and Fab198, especially in the CDR-H2 and CDR-H3 loops (See also Figure 4—figure supplement 1).