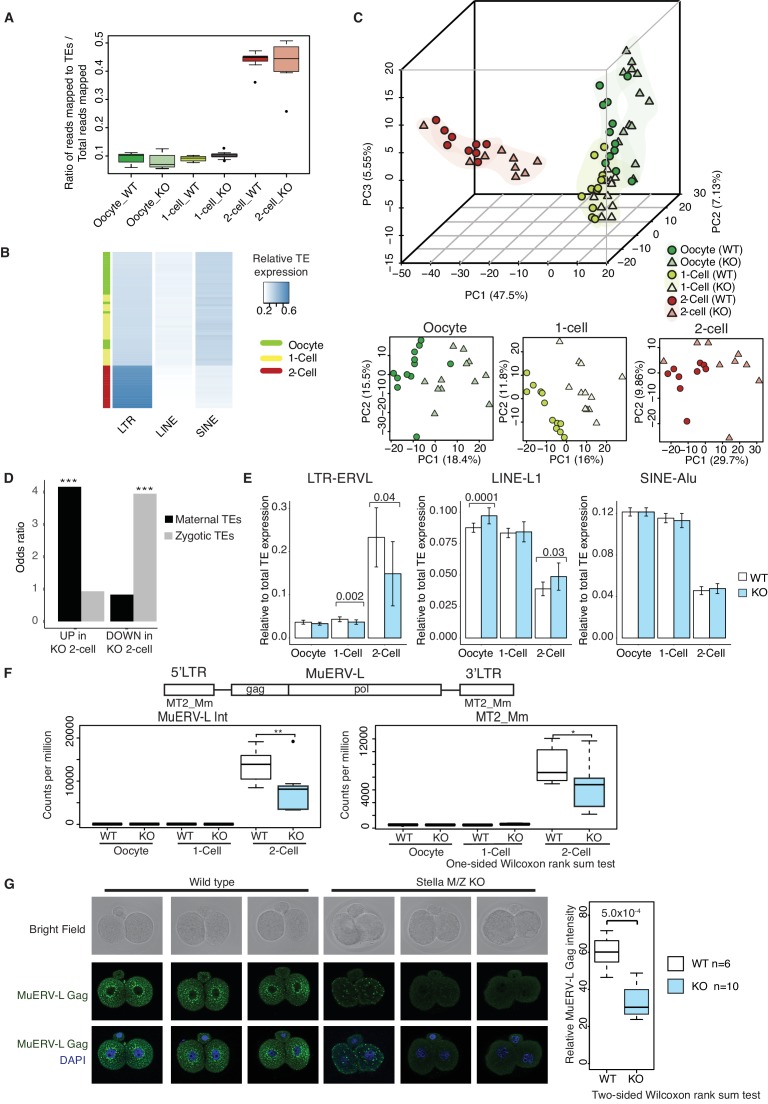
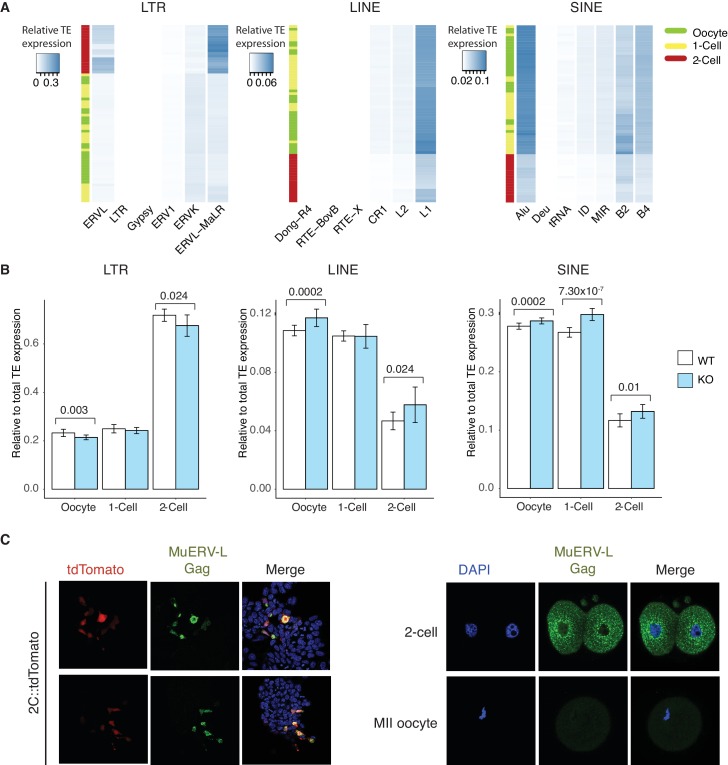
Figure 3. TEs are mis-expressed in the absence of Stella.
(A) Top panel shows boxplots of the ratio of reads mapped to TEs to total reads mapped in WT and KO oocytes, 1-cell and 2-cell embryos. The number of reads mapped to TEs are based on uniquely and multi-mapped counts (Figure 3—source data 2). (B) A heatmap of the relative expression of LTR, LINE and SINE in oocyte (green), 1-cell (yellow) and 2-cell embryos (red). Blue indicates higher expression and white indicates lower expression. Samples are clustered by row. (C) A score plot of the first three principal components for 66 cells using uniquely mapped TE counts (Figure 3—source data 1). The lower panels represent the score plots of the first two principal components using cells belonging to a specific developmental time point. The developmental time points are indicated by colour and the genotypes of Stella are indicated by shape. (D) A bar chart of the odds ratio of TEs up and downregulated in KO 2-cell embryos compared to WT, intersected with ‘maternal TEs’ and ‘zygotic TEs’. For maternal TEs enriched in TEs upregulated in KO 2-cell, ***p=5.35 × 10−6; for zygotic TEs enriched in TEs downregulated in KO 2-cell, ***p=8.13 × 10−7. (E) Bar plots showing the relative expression of TE families (LTR-ERVL, LINE1-L1 and SINE-Alu) in WT (white) and KO (light blue) oocytes, 1-cell and 2-cell embryos, data analysed from single-cell / embryo RNA sequencing. Two-sided Wilcoxon rank sum test performed between WT and KO samples and statistically significant p-values stated. (F) Top is an illustration of the structure of the full-length MuERV-L element flanked by 5’ and 3’ LTRs. Bottom is boxplots of the relative expression (counts per million) of MuERV-L Int and MT2_Mm transcript in WT (white) and KO (light-blue) oocytes, 1-cell and 2-cell embryos (p<0.05 corresponds to * and p<0.01 corresponds to **). (G) Immunofluorescence staining against MuERV-L Gag antibody in 2-cell embryos. (Left) The top panel shows bright-field, middle panel is MuERV-L Gag (green) and bottom panel are merged images of MuERV-L and DAPI, which counterstains DNA. Representative projections of z-stacks are shown for WT and KO embryos. (Right) A boxplot of the z-stack quantifications of the relative fluorescence intensity of MuERV-L Gag protein between WT and KO 2-cell embryos. Also see Figure 3—figure supplements 1–3 and Figure 3—source data 1–2.
DOI: http://dx.doi.org/10.7554/eLife.22345.014