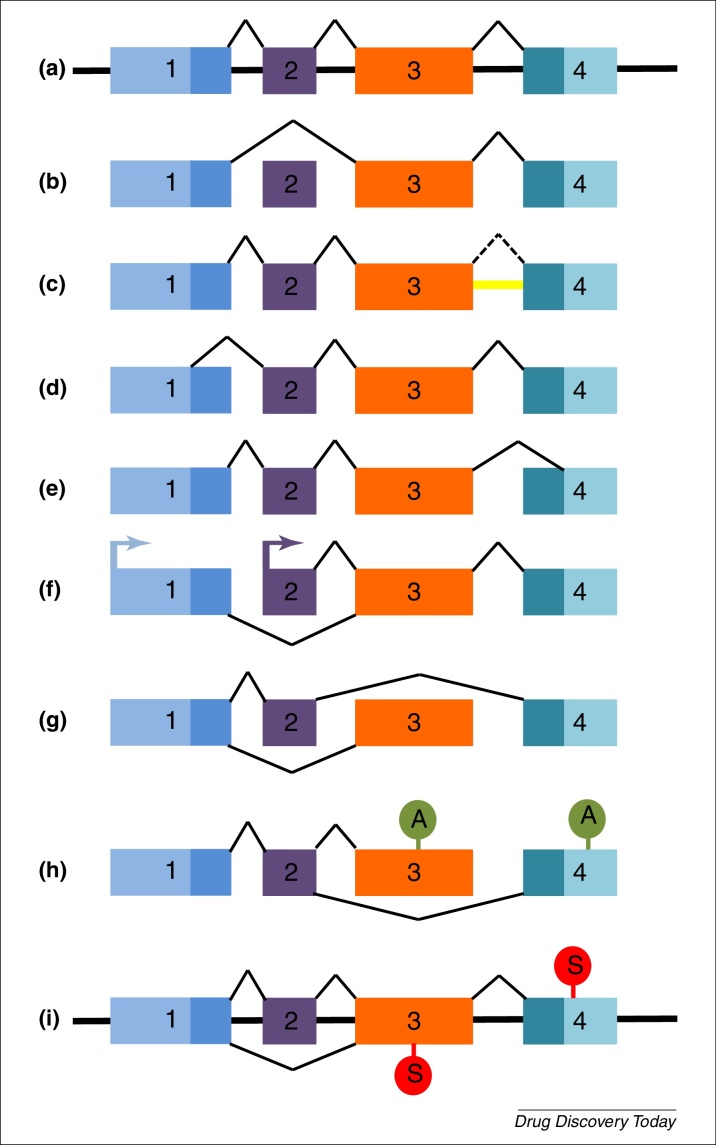
Figure 1.
Different types of alternative splicing compared with constitutive splicing. (a) Genomic structure showing the constitutive splicing of a pre-mRNA containing four exons, coloured boxes denote different exons. Alternative splice variants can arise by different mechanisms, such as: (b) exon skipping. Exon 2 is spliced out of the pre-mRNA; (c) intron retention. The intron between exons 3 and 4 (yellow) is not spliced out (dotted line) and is included in the mature mRNA; (d) alternative 5′ donor site. An intra-exonic splice site in exon 1 is used and gives an alternative 5′ sequence; (e) alternative 3′ acceptor site. An intra-exonic splice site is used in exon 4 and gives an alternative 3′ sequence; (f) alternative promoters; (g) mutually exclusive exons, where two mature mRNAs exist containing either exon 2 or exon 3 but never both exons together; (h) alternative polyadenylation sites; or (i) alternative stop codon use. Top stop codon: Constitutive splicing leads to use of stop codon in exon 4. Bottom stop codon: Exclusion of exon 2 leads to alternative stop codon in exon 3 and nonsense-mediated decay.