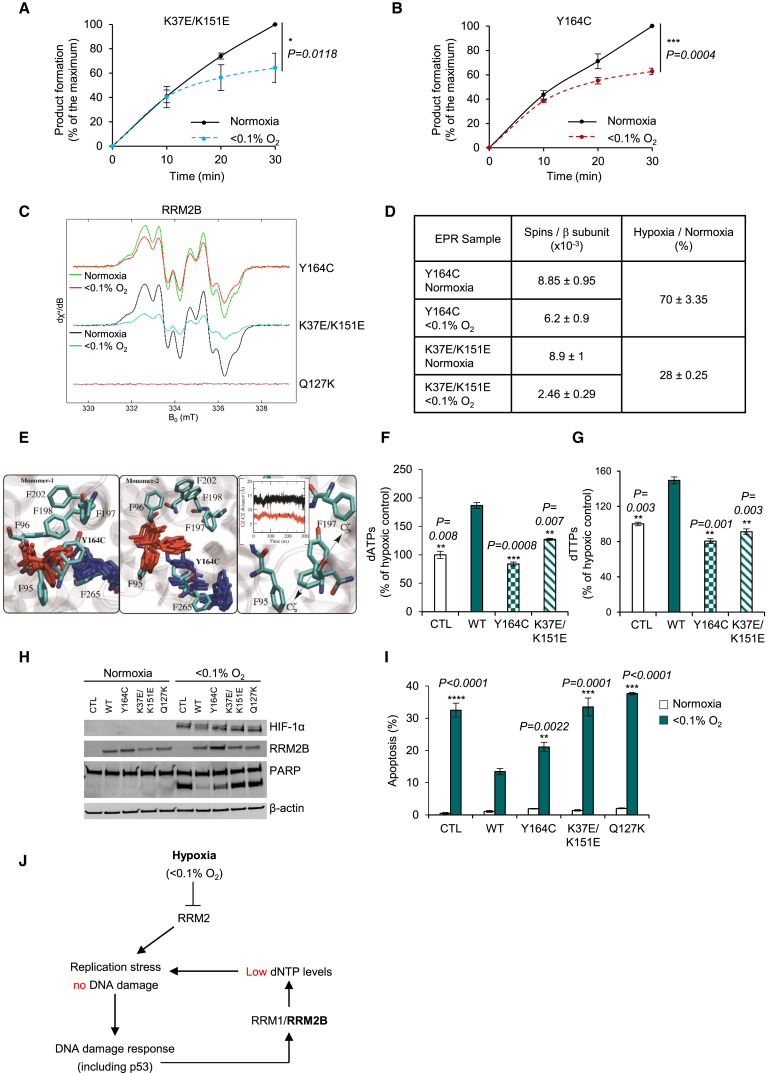
Figure 5.
Critical Roles of K37/K151 and Y164 in RRM2B
(A and B) Product formation (percentage of the maximum, where maximum is the dCDP levels at 30 min in normoxia) for K37E/K151E (A) and Y164C (B) in normoxia and <0.1% O2.
(C) EPR spectra of the tyrosyl radical of Y164C, K37E/K151E, and Q127K (as a negative control) in normoxia and <0.1% O2.
(D) Quantification of (C). Data present electron spins per β subunit.
(E) The RRM2B phenylalanine network around Y164 and phenylalanine conformation in Y164C mutation. Distance plot reveals the effect of Y164C in F95-F197 distance. Color code: WT (black), Y164C (red).
(F and G) dATP (F) and dTTP (G) levels in RKORRM2B−/− cells transfected with CTL, WT, Y164C, or K37E/K151E and exposed to <0.1% O2 (16 hr).
(H) Immunoblot for PARP cleavage in RKORRM2B−/− cells treated as in (F) and (G) plus Q127K and exposed to <0.1% O2 (19 hr).
(I) Apoptosis detected morphologically in RKORRM2B−/− cells treated as in (H).
(J) Schematic representation of our proposed model. Hypoxia leads to severely compromised activity of RRM2, leading to replication stress. RRM2B is then induced through the DDR pathway to maintain ongoing replication. However, insufficient dNTPs are generated by R1/R2B, and replication stress is unresolved. The importance of RRM2B activity is that while it does not resolve replication stress, it does maintain replication fork integrity and prevents the accumulation of DNA damage and loss of genome stability.
For (A), n = 3; for (B), n = 4 (biological replicates) and two-way ANOVA was applied; for (C), n = 2 (biological replicates); for (F)–(I), n = 3 (biological replicates); data represent means ± SEM and two-tailed Student’s t test was applied. See also Figures S6 and S7.