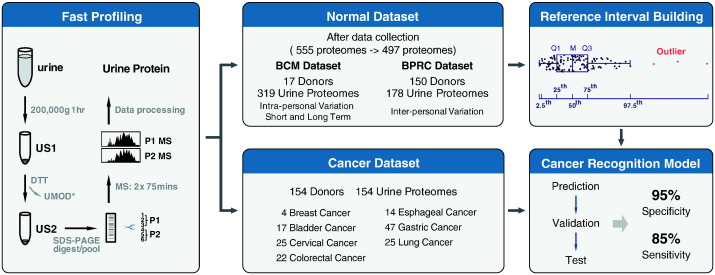
Fig. 1.
A schematic description of the workflow for fast profiling of urine proteome, establishing a reference interval (RI) of urine proteins for healthy human being and a cancer recognition algorithm. High-speed sediment of urine was collected by ultracentrifugation and uromodulin was removed by reduction with dithiotheitol. Then protein pellet was subjected to 1D SDS-PAGE for separation. After in-gel digestion, samples were pooled and measured by 2 MS runs. Normal dataset was acquired in an international two-center mode. Totally, 555 urine proteomes from 180 healthy donors were acquired. Of these data, 57 urine proteomes from 13 test subjects were collected during the method establishment phase in which centrifugation at low speed before ultracentrifugation or different temperatures in DTT reduction step for removing protein UMOD were tested to optimize our procedure. To keep technical consistency, these 57 urine proteomes were removed and only remaining 497 urine proteomes from 167 healthy donors were included in further analysis, with a BCM dataset (17 donors, 319 urine proteomes) and a BPRC dataset (150 donors, 178 urine proteomes). Personal and pan-human RI of urine proteomes were established with data in normal dataset. Cancer dataset (154 donors, 154 urine proteomes) included 7 types of solid tumors. Utilizing pan-human RI and data in cancer dataset, a cancer recognition algorithm was established to discriminate normal and cancer samples.