ABSTRACT
Molecular diagnosis of congenital toxoplasmosis or disseminated toxoplasmosis is based mainly on PCR. The repeated DNA element rep529 has become the main DNA target used in most PCR methods, whether laboratory developed or commercial. In this multicenter study, we evaluated the Toxoplasma ELITe MGB (Elitech) commercial kit by comparison with three reference quantitative PCR assays (RAs) used routinely in three proficient laboratories of the French National Reference Center for Toxoplasmosis network, using Toxoplasma calibrated suspensions diluted to obtain a range of concentrations from 0.1 to 10,000 parasites/ml. These suspensions were extracted with either the DNA extraction kit (EXTRAblood; Elitech) recommended by the manufacturer or the QIAamp DNA minikit (Qiagen). The Toxoplasma ELITe MGB assay was also evaluated on a panel of 128 clinical samples, including 56 amniotic fluid samples, 55 placenta samples, and various other samples, of which 95 originated from patients with proven toxoplasmosis. The ELITe MGB assay amplified low-concentration replicates (<10 parasites/ml) of calibrated suspensions less frequently than the RAs of 2/3 laboratories. Additionally, the combination of EXTRAblood and Toxoplasma ELITe MGB yielded poorer sensitivity than the combination of QIAamp DNA minikit and ELITe MGB for low parasite concentrations (P < 0.001 for 1 parasite/ml). On clinical samples, the sensitivity and the specificity of the commercial assay were 89% and 100%, respectively. The sensitivity ranged from 79% (placenta samples) to 100% (amniotic fluid samples). Overall, this study shows that the Toxoplasma ELITe MGB assay is suitable for the diagnosis of toxoplasmosis from non-cell-rich or non-hemoglobin-rich samples and that the EXTRAblood kit is not optimal.
KEYWORDS: PCR, Toxoplasma gondii, diagnosis, rep529, toxoplasmosis
INTRODUCTION
Toxoplasmosis is a worldwide parasitic disease caused by the intracellular coccidian parasite Toxoplasma gondii. Molecular diagnosis is an essential tool for the diagnosis of congenital toxoplasmosis as well as acute disease in immunocompromised patients with primary infection or reactivation of past infection (1). Prenatal diagnosis of congenital toxoplasmosis relies on Toxoplasma DNA detection in amniotic fluid (AF) and has been largely evaluated in field studies, particularly in French series, as a national prevention program implemented in 1992 requires a monthly serological follow-up of seronegative pregnant women and recommends the use of amniocentesis when a primary infection is documented. In France, 21 university hospitals have a ministerial agreement for the prenatal diagnosis of congenital toxoplasmosis. Over the last several years, all of them have moved to the use of real-time PCR (RT-PCR) methods targeting the repeated DNA element rep529 (GenBank accession number AF146527), as the sensitivity provided using this DNA target has proved to be higher than that of the formerly used B1 gene in most studies (2–5). About three-quarters of these reference laboratories still use “in-house” or laboratory-developed RT-PCR techniques which have been evaluated in clinical studies, but there is an increasing trend to use commercial assays, which, in spite of being more expensive, are easier to use and allow better quality management than the former. The manufacturers of these kits announce a sensitivity threshold, but the performance of these assays may be altered by the tested sample type or the DNA extraction method used. Indeed, Toxoplasma may be searched not only for the prenatal diagnosis of congenital toxoplasmosis but also in other sample types from immunocompromised patients, such as blood, bronchoalveolar lavage fluid, cerebrospinal fluid (CSF), aqueous humor (AH), or various biopsy specimens (6). In view of the globally excellent performances of “in-house” methods in proficient diagnostic centers (7), the evaluation of the analytical and clinical performances of commercial kits is absolutely needed to ensure the quality of results in routine use. Moreover, some commercial assays are validated for Toxoplasma detection in AF but not in other sample types. In this multicenter study, we evaluated the performances of the Toxoplasma ELITe MGB kit (Elitech, Puteaux, France), a rep529-targeting assay, using serial dilutions of calibrated Toxoplasma suspensions in AF and using clinical samples, including AF, placenta, and various other types of samples.
RESULTS
Comparative testing using Toxoplasma calibrated suspensions.
The first step of the study was to determine the PCR performance scores using serial dilutions of calibrated Toxoplasma DNA suspensions. PCR performance scores were calculated as described elsewhere (7, 8), using the T. gondii DNA serial dilution assay, and are reported in Table 1. In all three centers, the scores obtained using the in-house method and the Elitech assay were close, but the reference assays (RA) method had a higher score than the commercial assay in two out of three centers (P < 0.01) (Table 1). Taken together, the lowest parasite concentrations, i.e., “1” and “0.1” Toxoplasma parasites/ml, were inconstantly amplified. For 1 parasite/ml, the difference was statistically significant (5 positive out of 10 replicates and 8 positive out of 10 replicates with ELITe MGB and RAs, respectively) (P < 0.01).
TABLE 1.
Performance scores for the four PCR assays using DNA serial dilutions of a calibrated Toxoplasma suspensiona
| Concn (T. gondii parasites/ml) | No. of positive reactions/no. of reactions performed |
||||||
|---|---|---|---|---|---|---|---|
| Lab 1 |
Lab 2 |
Lab 3 |
|||||
| RA1 (Qiagen) | ELITe MGB (Qiagen) | ELITe MGB (EXTRAblood + IC) | RA2 (Qiagen) | ELITe MGB (Qiagen) | RA3 (Qiagen) | ELITe MGB (Qiagen) | |
| 10,000 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 4/4 | 4/4 |
| 1,000 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 4/4 | 4/4 |
| 100 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 4/4 | 4/4 |
| 10 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 4/4 | 4/4 |
| 1 | 3/3 | 2/3 | 2/3 | 1/3 | 0/3 | 4/4 | 3/4 |
| 0.1 | 0/3 | 0/3 | 0/3 | 0/3 | 1/3 | 1/4 | 1/4 |
| PCR performance score (%) | 15/18* (83.3) | 14/18 (77.8) | 14/18 (77.8) | 13/18 (72.2) | 13/18 (72.2) | 21/24* (87.5) | 20/24 (83.3) |
RA, reference assay. *, P < 0.01 compared to ELITe MGB/Qiagen.
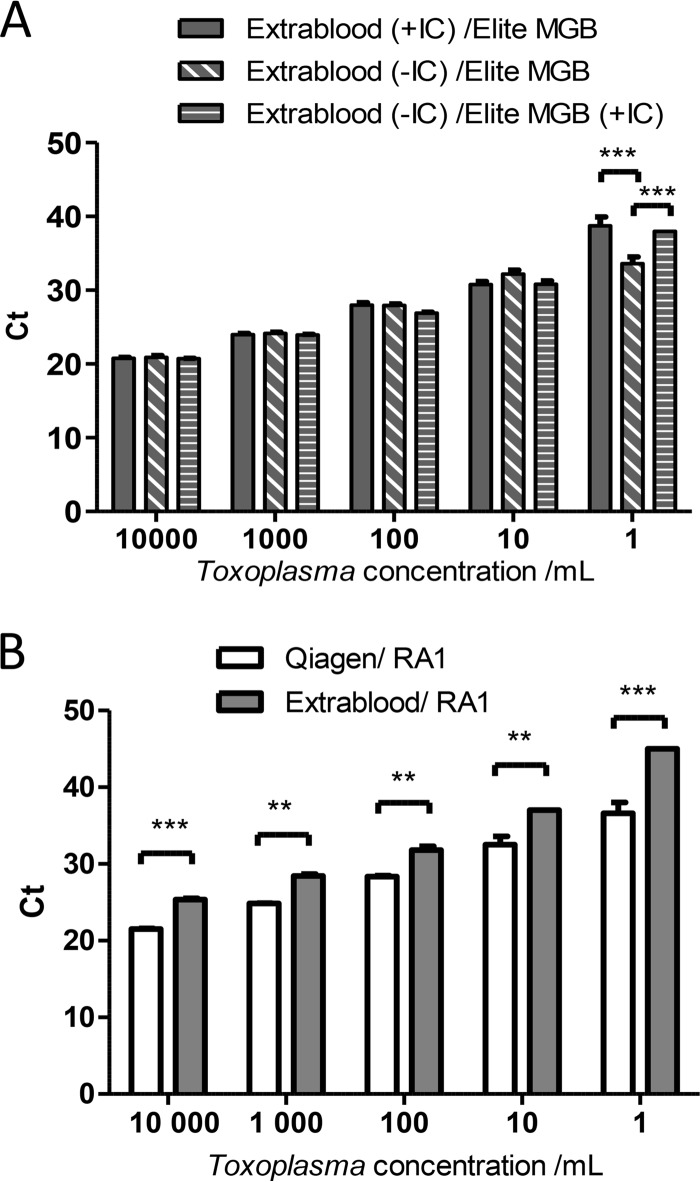
The mean cycle threshold (CT) of amplification obtained with Toxoplasma ELITe MGB was calculated for concentrations of calibrated suspensions between 1 and 10,000 T. gondii parasites/ml, and the results were compared for the two DNA extraction methods. Surprisingly, the mean CTs obtained with the combination EXTRAblood/ELITe MGB (recommended by the manufacturer) were significantly higher than those obtained with the combination QIAamp/ELITe MGB (29.2 ± 0.17 versus 27.8 ± 0.18 [P < 0.05] for 100 T. gondii parasites/ml and 39.3 ± 0.07 versus 34.6 ± 0.4 [P < 0.001] for 1 T. gondii parasite/ml) (Fig. 1).
FIG 1.
Evaluation of the Toxoplasma ELITe MGB kit using serial dilutions of Toxoplasma calibrated suspensions. Calibrated suspensions were extracted using either the QIAamp DNA minikit or EXTRAblood before being amplified using Toxoplasma ELITe MGB. Amplifications were performed in triplicate. Data are expressed as means ± standard errors of the means (SEM). *, P < 0.05; ***, P < 0.001.
Comparative testing using clinical samples.
When using clinical samples, the concordance between the in-house method and the ELITe MGB assay was 92% (118/128). All negative DNAs tested negative (33/33), but false-negative results were obtained using the ELITe MGB kit compared with the RAs for 10 out of 95 positive samples, yielding a specificity of 100% and a relative sensitivity of 89.5% for the kit. This sensitivity was 100% for AF (38/38) and other fluid samples (AH and CSF) but only 79% (34/43) for placenta and 80% (4/5) for buffy coat samples (Table 2). In 9/10 cases, the false-negative results were obtained for placenta samples with high CT values (>37). To rule out PCR inhibition, samples were diluted 1/10 and retested. Additionally, as the nature of the Elitech internal control (IC) was not known and could be suspected to interfere with small amounts of parasite DNA, these false-negative samples were also tested without the IC. Amplification was restored from plain DNA in the absence of IC in two cases (with CTs of 39.7 and 40.7) and after dilution to 1/10 and in the absence of IC in one case (CT = 36.7). No amplification was observed after dilution to 1/10 when the IC was not removed.
TABLE 2.
Performance of the Toxoplasma ELITe MGB assay using clinical samples
| Sample type (n) | Sensitivity, % (n/N) | Specificity, % (n/N) |
|---|---|---|
| Clinical samples (128) | 89.5 (85/95) | 100 (33/33) |
| Amniotic fluid (56) | 100 (38/38) | 100 (18/18) |
| Other fluidsa (9) | 100 (7/7) | 100 (2/2) |
| Placenta (55) | 79 (34/43) | 100 (12/12) |
| Blood (buffy coat) (6) | 80 (4/5) | 100 (1/1) |
| Biopsy specimens (2) | 100 (2/2) | NAb |
| QCMD samples (10) | 100 (7/7) | 100 (3/3) |
Five cerebrospinal fluid samples and 4 aqueous humor samples.
NA, not applicable.
Overall, the sensitivity for fluid samples was higher than that for cellular samples (100% versus 80%; P < 0.01) (Table 2).
Finally, the qualitative results obtained using the QCMD 2014 quality control (QC) samples were concordant for the RA1 and ELITe MGB assays (7/7 positive samples and 3/3 negative samples) (Table 2). Mean CTs obtained with the two techniques were not statistically different (P = 0.8) (data not shown).
Influence of IC on amplification performances.
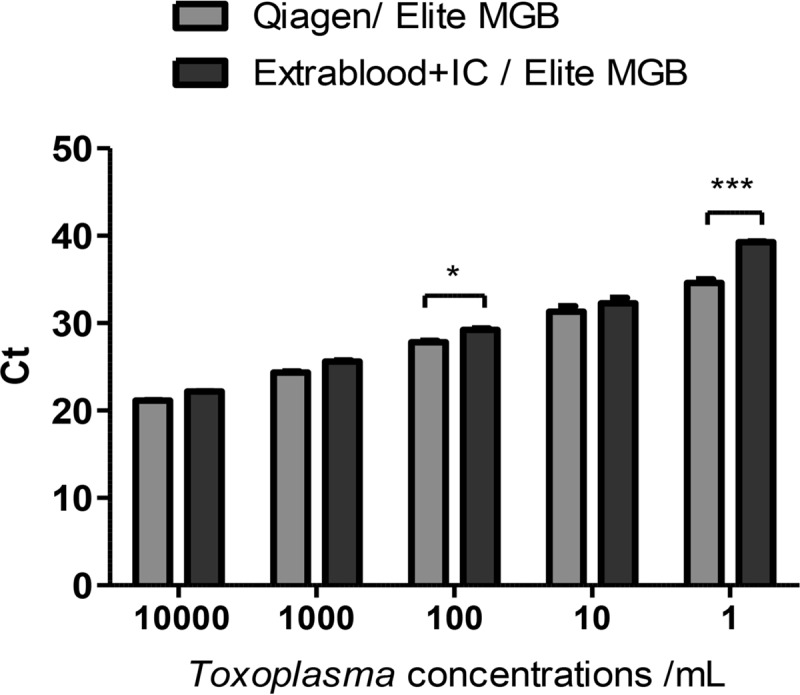
As the observation above suggested a competing effect of the IC, another series of DNA extractions of a calibrated Toxoplasma suspension using different conditions was launched to confirm this hypothesis. It appeared that DNA extraction with EXTRAblood (with 5 μl of IC added in the sample as recommended) followed by amplification with ELITe MGB performed as well as extraction with EXTRAblood followed by addition of IC at the time of amplification with ELITe MGB (Fig. 2A). Surprisingly, at the “1 T. gondii parasite/ml” concentration, the mean CT was significantly lower when the IC was withdrawn from the extraction and amplification steps than that obtained after extraction with EXTRAblood including IC (33.57 ± 0.93 versus 38.75 ± 1.18; P < 0.001) (Fig. 2A). Similarly, the addition of the IC to the amplification mix also led to an increase of the mean CT at the “1 T. gondii parasite/ml” concentration (33.57 ± 0.93 versus 37.97 ± 0.007; P < 0.001). In another experiment, the extraction methods (QIAamp DNA minikit and EXTRAblood) were compared using the same amplification method (RA1). Whatever the parasite concentration, the CT values were much lower when DNA was extracted with the QIAamp DNA minikit than with EXTRAblood (36.62 ± 1.38 versus 45 and 21.52 ± 0.08 versus 25.35 ± 0.2 for the lowest and highest parasite concentrations, respectively; P < 0. 001) (Fig. 2B).
FIG 2.
Evaluation of the impact of the internal control (IC) and of the extraction method on amplification efficacy. (A) Comparison of the CTs of amplification obtained with ELITe MGB PCR on DNA extracted with EXTRAblood, with addition of the internal control (+IC) in the sample before extraction or without the internal control (−IC) or after addition of IC in DNA at the time of amplification. (B) Comparison of the CTs obtained with reference assay 1 (RA1) after DNA extraction with EXTRAblood or with the QIAamp DNA minikit. Amplifications were performed in triplicate; data are expressed as means ± SEM. **, P < 0.01; ***, P < 0.001.
DISCUSSION
In the diagnosis of congenital toxoplasmosis, a high sensitivity of the PCR assay is needed, as parasite loads in amniotic fluids from congenitally infected infants are frequently as low as 10 tachyzoites/ml or less (9). Early treatment of pregnant women who benefit from serological screening in France could account for such low parasite loads (10). High performance of molecular assays is also a key issue in immunocompromised patients, for whom rapid and accurate diagnosis is essential. Furthermore, toxoplasmosis is of increasing importance in HIV-negative immunocompromised patients, due to the growing number of transplantations and to the use of immunosuppressive drugs for the treatment of chronic inflammatory diseases (6). Here, when applied to calibrated DNA samples, the Toxoplasma ELITe MGB assay showed performance similar to those of the three laboratory-developed reference methods down to 10 T. gondii parasites/ml, but the lowest concentrations were inconstantly detected (Table 1), despite a higher DNA input into the amplification reaction mixture (10 μl) than in the reference methods (Table 3).
TABLE 3.
Technical description of the four PCR methods useda
| Technique | DNA target | Type of probe | DNA input (μl) | Internal control | No. of cycles | PCR device |
|---|---|---|---|---|---|---|
| RA1 | rep529 | TaqMan (FAM-TAMRA) | 5 | Universal extraction and inhibition DNA control (Diagenode) | 40 | StepOnePlus (ThermoFisher) |
| RA2 | rep529 | TaqMan | 5 | Noncompetitive exogenous DNA inserted in PCR 2.1 vector | 50 | AB7000 (ThermoFisher) |
| RA3 | rep529 | FRET | 7 | PhiX (DNA bacteriophage) | 40 | LC2.0 (ThermoFisher) |
| Toxoplasma ELITe MGB | rep529 | TaqMan (FAM-MGB) | 10 | Artificial DNA sequence | 45 | StepOnePlus (ThermoFisher) |
RA, reference assay; FAM, 6-carboxyfluorescein; TAMRA, 6-carboxytetramethylrhodamine; FRET, fluorescence resonance energy transfer.
When using clinical samples, the Toxoplasma ELITe MGB assay showed 100% sensitivity in amniotic fluids and yielded congruent results on a batch of external quality controls (QCMD 2014). Although the kit is commercialized, i.e., validated, only for amniotic fluids and whole blood samples, we included other sample types which are frequently sent to laboratories for diagnosis, such as aqueous humor or CSF. We also included placenta samples, which offer the advantage of mimicking blood samples and being linked to a clinical history and follow-up of neonates and thus can be definitely classified as true positive or not. The sensitivity for other fluid samples was as good as that for AF, although the number of samples included was small. However, the sensitivity was much lower for other sample types, in particular placenta samples. These results show that this commercial assay performs better on samples with low cellularity or reduced contamination with red blood cells, which confirms the lower sensitivity using whole blood reported by the manufacturer itself. As routinely done in the three reference laboratories, when PCR inhibition was suspected from the observation of high CT values with their IC, samples were retested after sample dilution. This strategy was thus applied to the false-negative samples obtained with the ELITe MGB assay, but it did not provide amplification restoration. Additionally, as the nature of the Elitech IC was not known and could be suspected to interfere with small amounts of parasite DNA, these false-negative samples were also tested without the IC. This allowed restoration of Toxoplasma amplification in 30% of these samples, thus demonstrating that competition of the Elitech IC could partially explain the false-negative results. The inhibitory effect of the Elitech IC was definitely confirmed using low concentrations of Toxoplasma calibrated suspensions (Fig. 2A). However, the technical instructions from the manufacturer do not mention that the reactions should be performed in duplicate, plain and diluted, or with and without the IC, as this significantly augments the cost of the test. With respect to the interpretation of IC amplification, it is stated in the manufacturer's instructions that a CT value of ≤35 is suitable for a correct interpretation of results. During our assays, the CT value obtained for the IC was always <30 and thus it could be reasonably considered that the test was valid, yet false-negative results have been found.
As the DNA extraction method is also known to influence the performance of PCR amplification (11), we decided to compare the CT results for calibrated parasite suspensions, extracted using either the extraction kit (EXTRAblood) as recommended by the manufacturer or the QIAamp DNA blood minikit, as routinely done in the three labs. Subsequent amplification with RA1 showed that the combination of QIAamp extraction and RA1 performed much better than the combination of EXTRAblood and RA1 (Fig. 2B), stressing the need to evaluate the extraction and PCR methods together, as emphasized earlier (12). Moreover, the combination of EXTRAblood and ELITe MGB (recommended by the manufacturer) performed significantly worse than the combination of QIAamp and ELITe MGB to amplify low parasite concentrations (Fig. 1) and thus should be avoided. The evaluation of the combination of extraction and amplification methods is of peculiar importance for off-label use of commercial PCR assays, particularly with cell-rich samples types, and its capacity to detect very small parasite amounts must be verified. Furthermore, the validation of the entire process is necessary to meet the requirements of the quality assurance system.
Regarding convenience and good laboratory practices, the mix provided in the kit contains uracil-N-glycosylase (UNG) to limit carryover contaminations from previously amplified PCR products. The kit does not include a standard to determine a curve to quantify parasite loads. The PCR assay, the internal control, and the positive control are purchased in three separate vials; thus, the management of batch traceability is not easy.
Several quantitative PCR (qPCR) commercial assays are currently available to detect Toxoplasma DNA, but very few have been evaluated using clinical samples or in routine use. A previous multicenter study conducted by the Molecular Biology Group of the French National Reference Center for Toxoplasmosis (NRCT) evaluated another commercial kit by Bio-Evolution using 157 amniotic fluid samples and found a 99% concordance for T. gondii-infected samples (13). In the present study, we included fewer amniotic fluid samples (56), but the Toxoplasma ELITe MGB assay also showed a good concordance using these samples.
Taking the results together, this study showed that the Toxoplasma ELITe MGB assay appears to be suitable for prenatal diagnosis. However, a note of caution is in order when using cell-rich or hemoglobin-rich samples, as this carries the risk of false-negative results. In our hands, the use of the EXTRAblood DNA extraction lowered the performance of the Toxoplasma ELITe MGB assay for low parasite concentrations and dramatically lowered the sensitivity of the in-house PCR method tested. Like for all molecular diagnostic methods, clinical microbiologists who would aim at implementing this technique in their laboratory should evaluate the combination of extraction and amplification methods before changing one or the other.
MATERIALS AND METHODS
Participating centers.
The three participating centers (university hospitals of Paris-Cochin, Rennes, and Grenoble, France) are part of the Molecular Biology Study Group of the French National Reference Center for Toxoplasmosis (NRCT) http://cnrtoxoplasmose.chu-reims.fr/?lang=en) and have a ministerial agreement for the prenatal diagnosis of toxoplasmosis. They participate as national external quality controls on a regular basis with satisfactory results.
Samples. (i) Calibrated Toxoplasma suspensions.
The three participating centers used a calibrated Toxoplasma (type II) suspension produced by the Molecular Biology Study Group of the NRCT (University Hospital of Montpellier) (14). DNA was previously extracted from the suspension in each center using their routine method for molecular diagnosis (QIAamp DNA minikit; Qiagen, Les Ulis, France). The three labs compared the sensitivities of detection of serial dilutions (from 10,000 tachyzoites/ml to 0.1/ml) after amplification using their own in-house PCR method (so-called reference assays [RAs]) (Table 3) and the Toxoplasma ELITe MGB kit, following the manufacturer's instructions. All dilution points were amplified in triplicate or quadruplicate.
Another batch of calibrated suspension was extracted in parallel, using either the manual extraction device recommended by the manufacturer, i.e., EXTRAblood (Elitech), or the QIAamp DNA minikit. In this experiment, the internal control was added at the time of extraction, following the manufacturer's instructions (see below).
Additionally, one center also evaluated a batch of 10 samples of external quality controls (EQC) from QCMD (2014). This QC batch (TGDNA 14) consisted of 5 vials of lyophilized amniotic fluid samples spiked with various concentrations of T. gondii or unspiked (negative) and 5 vials of lyophilized plasma samples spiked with various concentrations of T. gondii or unspiked (negative) and was extracted using EXTRAblood (Elitech).
(ii) Clinical samples.
This part of the work used stored DNA from clinical samples obtained during routine molecular diagnosis (2005 to 2015). The three reference laboratories selected Toxoplasma-positive and -negative DNAs preserved at −20°C or −80°C following routine molecular diagnosis (15). Because of the French prevention program for congenital toxoplasmosis, reference centers have a collection of samples (mainly AF and placenta samples) from patients with confirmed diagnosis. As already evaluated in previous studies, long-term storage of DNA at −20°C or below does not alter the result of Toxoplasma real-time PCR (3, 15), thus allowing the use of collections for diagnostic evaluations. Placenta and AF DNA samples from fetuses with suspected congenital toxoplasmosis were classified as true positive or true negative on the basis of the newborn serological follow-up (detection of specific IgM or IgA and comparison of mother and newborn antibody profiles by Western blotting during a 1-year follow-up) so that the diagnosis could be definitely confirmed or ruled out, as previously described (13, 16–18). The Elitech assay was certified CE-IVD in 2013 and validated for amniotic fluid samples and whole blood, and thus we considered that placenta samples were comparable to blood samples. Other samples were collected from patients with retinochoroiditis or from immunocompromised patients, for whom the clinical diagnosis was recorded (disseminated, cerebral, or toxoplasmosis excluded). Overall, 128 DNA samples were included: 55 isolated from placentas, 56 from AF, 4 from AH, 6 leukocyte pellets isolated from buffy coats, 2 from biopsy specimens, and 5 from CSF. Of these, 33 samples were classified as PCR negative and 95 as PCR positive, according to serological and clinical follow-up of patients.
Molecular techniques. (i) DNA extraction of clinical specimens.
Clinical specimens were processed in the setting of routine diagnosis. After appropriate preanalytical steps (centrifugation of fluids, buffy coat, and predigestion of placenta or biopsy specimens with proteinase K), 200 μl of clinical samples was extracted using the QIAamp DNA minikit according to the manufacturer's instructions and eluted in 100 μl (all three centers used the same technique for routine diagnosis).
(ii) DNA extraction of Toxoplasma calibrated suspensions.
In each center, the same batch of Toxoplasma calibrated suspension was extracted using the QIAamp DNA minikit and amplified in parallel with the RA and the ELITe MGB assay. In one center, one batch of calibrated suspension was also extracted using EXTRAblood, with or without adding 5 μl of internal control (IC) (CPE-DNA internal control; Elitech), following the manufacturer's instructions. Briefly, 200 μl of samples was mixed with 25 μl of proteinase K, 200 μl of lysis buffer, 10 μl of carrier RNA, and 5 μl of IC, incubated at 70°C for 10 min, and then centrifuged for 5 s at 11,000 rpm. After addition of 210 μl of absolute ethanol and a brief centrifugation, the lysate was loaded into a column. After several washing steps, DNA was eluted in 60 μl of buffer. Quality control samples (QCMD 2014) were also extracted using this technique.
(iii) DNA amplification. (a) Reference methods.
The in-house methods used by the three participants targeted the rep529 sequence and have been previously evaluated and published (5, 14, 16, 19). Technical details can be found in Table 3. All three RAs satisfy the annual external quality control program managed by the National Reference Center for Toxoplasmosis (Centre Hospitalier Universitaire de Montpellier). All DNAs from clinical samples were reamplified in parallel with the RA method and the ELITe MGB assay. Newly extracted Toxoplasma suspensions were amplified with both techniques.
(b) Toxoplasma ELITe MGB method.
The Toxoplasma ELITe MGB assay was performed according to the manufacturer's instructions, using 10 μl of the DNA template and 20 μl of mix. When the Elitech IC was not added during extraction (calibrated suspensions or clinical samples previously extracted using the QIAamp DNA minikit), 2 μl of Elitech IC was added in 10 μl of template DNA, and 10 μl of this solution was used for amplification, as suggested by the manufacturer.
The assay has been validated only on Applied Biosystems devices. In this study, ELITe MGB amplification was performed using a StepOnePlus device or ABI Prism 7000 (ThermoFisher) and the following program: 2 min at 50°C, 2 min at 94°C, and 45 cycles of 10 s at 94°C, 30 s at 60°C, and 20 s at 72°C. Each clinical sample was analyzed in a single reaction, as well as the QCMD EQC DNA. If the result was not concordant with that of the previous routine diagnosis, a second PCR was performed with the Elitech method and the in-house PCR. The limits of detection (LODs) (95% sensitivity) announced by the manufacturer for whole blood samples are 34.29 or 88.72 T. gondii parasites/ml and 5.47 or 1.91 T. gondii parasites/extraction in AF samples, using manual extraction (EXTRAblood; Elitech) or automated extraction (NucliSENS EasyMAG; bioMérieux), respectively.
(c) Performance score.
For each technique, a performance score was calculated from the results obtained with calibrated suspensions, as follows: number of positive replicates/total number of amplifications.
Statistical analysis.
The Mann-Whitney test was used to compare the mean CTs obtained with qPCR assays for the calibrated Toxoplasma suspensions. When the number of positive replicates was below three, a two-way analysis of variance (ANOVA) was used instead.
For clinical samples, the analysis of qualitative results obtained with both techniques was analyzed using a chi-square test or a Fisher exact test.
Statistical analysis was done using GraphPad Prism V5 (GraphPad Software, USA). A P value of <0.05 was considered significant.
ACKNOWLEDGMENTS
This study was in part funded by the Institut de Veille Sanitaire (InVS) through financial support of the Molecular Biology Study Group of the French National Reference Centre for Toxoplasmosis (Centre National de Référence de la Toxoplasmose).
The members of the Molecular Biology Study Group of the French National Reference Center for Toxoplasmosis are Patrick Bastien (University Hospital of Montpellier), Marie-Pierre Brenier-Pinchart (University Hospital of Grenoble), Sophie Cassaing (University Hospital of Toulouse), Frédéric Dalle (University Hospital of Dijon), Laurence Delhaes (University Hospital of Bordeaux), Denis Filisetti (University Hospital of Strasbourg), Jean Ménotti (University Hospital of Saint-Louis, Paris), Hervé Pelloux (University Hospital of Grenoble), Florence Robert-Gangneux (University Hospital of Rennes), Yvon Sterkers (University Hospital of Montpellier), Feriel Touafek (University Hospital of Pitié-Salpêtrière, Paris), Emmanuelle Varlet-Marie (University Hospital of Montpellier), and Hélène Yera (University Hospital of Cochin, Paris).
REFERENCES
- 1.Robert-Gangneux F, Darde ML. 2012. Epidemiology of and diagnostic strategies for toxoplasmosis. Clin Microbiol Rev 25:264–296. doi: 10.1128/CMR.05013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cassaing S, Bessieres MH, Berry A, Berrebi A, Fabre R, Magnaval JF. 2006. Comparison between two amplification sets for molecular diagnosis of toxoplasmosis by real-time PCR. J Clin Microbiol 44:720–724. doi: 10.1128/JCM.44.3.720-724.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belaz S, Gangneux JP, Dupretz P, Guiguen C, Robert-Gangneux F. 2015. A 10-year retrospective comparison of two target sequences, REP-529 and B1, for Toxoplasma gondii detection by quantitative PCR. J Clin Microbiol 53:1294–1300. doi: 10.1128/JCM.02900-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kasper DC, Sadeghi K, Prusa AR, Reischer GH, Kratochwill K, Forster-Waldl E, Gerstl N, Hayde M, Pollak A, Herkner KR. 2009. Quantitative real-time polymerase chain reaction for the accurate detection of Toxoplasma gondii in amniotic fluid. Diagn Microbiol Infect Dis 63:10–15. doi: 10.1016/j.diagmicrobio.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 5.Reischl U, Bretagne S, Kruger D, Ernault P, Costa JM. 2003. Comparison of two DNA targets for the diagnosis of toxoplasmosis by real-time PCR using fluorescence resonance energy transfer hybridization probes. BMC Infect Dis 3:7. doi: 10.1186/1471-2334-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robert-Gangneux F, Sterkers Y, Yera H, Accoceberry I, Menotti J, Cassaing S, Brenier-Pinchart MP, Hennequin C, Delhaes L, Bonhomme J, Villena I, Scherer E, Dalle F, Touafek F, Filisetti D, Varlet-Marie E, Pelloux H, Bastien P. 2015. Molecular diagnosis of toxoplasmosis in immunocompromised patients: a 3-year multicenter retrospective study. J Clin Microbiol 53:1677–1684. doi: 10.1128/JCM.03282-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sterkers Y, Varlet-Marie E, Cassaing S, Brenier-Pinchart MP, Brun S, Dalle F, Delhaes L, Filisetti D, Pelloux H, Yera H, Bastien P. 2010. Multicentric comparative analytical performance study for molecular detection of low amounts of Toxoplasma gondii from simulated specimens. J Clin Microbiol 48:3216–3222. doi: 10.1128/JCM.02500-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morelle C, Varlet-Marie E, Brenier-Pinchart MP, Cassaing S, Pelloux H, Bastien P, Sterkers Y. 2012. Comparative assessment of a commercial kit and two laboratory-developed PCR assays for molecular diagnosis of congenital toxoplasmosis. J Clin Microbiol 50:3977–3982. doi: 10.1128/JCM.01959-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Romand S, Chosson M, Franck J, Wallon M, Kieffer F, Kaiser K, Dumon H, Peyron F, Thulliez P, Picot S. 2004. Usefulness of quantitative polymerase chain reaction in amniotic fluid as early prognostic marker of fetal infection with Toxoplasma gondii. Am J Obstet Gynecol 190:797–802. doi: 10.1016/j.ajog.2003.09.039. [DOI] [PubMed] [Google Scholar]
- 10.Wallon M, Peyron F, Cornu C, Vinault S, Abrahamowicz M, Kopp CB, Binquet C. 2013. Congenital toxoplasma infection: monthly prenatal screening decreases transmission rate and improves clinical outcome at age 3 years. Clin Infect Dis 56:1223–1231. doi: 10.1093/cid/cit032. [DOI] [PubMed] [Google Scholar]
- 11.Yera H, Filisetti D, Bastien P, Ancelle T, Thulliez P, Delhaes L. 2009. Multicenter comparative evaluation of five commercial methods for toxoplasma DNA extraction from amniotic fluid. J Clin Microbiol 47:3881–3886. doi: 10.1128/JCM.01164-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bastien P, Procop GW, Reischl U. 2008. Quantitative real-time PCR is not more sensitive than “conventional” PCR. J Clin Microbiol 46:1897–1900. doi: 10.1128/JCM.02258-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Filisetti D, Sterkers Y, Brenier-Pinchart MP, Cassaing S, Dalle F, Delhaes L, Pelloux H, Touafek F, Varlet-Marie E, Yera H, Candolfi E, Bastien P. 2015. Multicentric comparative assessment of the bio-evolution Toxoplasma gondii detection kit with eight laboratory-developed PCR assays for molecular diagnosis of congenital toxoplasmosis. J Clin Microbiol 53:29–34. doi: 10.1128/JCM.01913-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Varlet-Marie E, Sterkers Y, Brenier-Pinchart MP, Cassaing S, Dalle F, Delhaes L, Filisetti D, Pelloux H, Touafek F, Yera H, Bastien P. 2014. Characterization and multicentric validation of a common standard for Toxoplasma gondii detection using nucleic acid amplification assays. J Clin Microbiol 52:3952–3959. doi: 10.1128/JCM.01906-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delhaes L, Filisetti D, Brenier-Pinchart MP, Pelloux H, Yera H, Dalle F, Sterkers Y, Varlet-Marie E, Touafek F, Cassaing S, Bastien P. 2014. Freezing and storage at −20 degrees C provides adequate preservation of Toxoplasma gondii DNA for retrospective molecular analysis. Diagn Microbiol Infect Dis 80:197–199. doi: 10.1016/j.diagmicrobio.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 16.Robert-Gangneux F, Dupretz P, Yvenou C, Quinio D, Poulain P, Guiguen C, Gangneux JP. 2010. Clinical relevance of placenta examination for the diagnosis of congenital toxoplasmosis. Pediatr Infect Dis J 29:33–38. doi: 10.1097/INF.0b013e3181b20ed1. [DOI] [PubMed] [Google Scholar]
- 17.Delhaes L, Yera H, Ache S, Tsatsaris V, Houfflin-Debarge V. 2013. Contribution of molecular diagnosis to congenital toxoplasmosis. Diagn Microbiol Infect Dis 76:244–247. doi: 10.1016/j.diagmicrobio.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 18.Filisetti D, Yera H, Villard O, Escande B, Wafo E, Houfflin-Debarge V, Delhaes L, Bastien P. 2015. Contribution of neonatal amniotic fluid testing to diagnosis of congenital toxoplasmosis. J Clin Microbiol 53:1719–1721. doi: 10.1128/JCM.02358-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Talabani H, Asseraf M, Yera H, Delair E, Ancelle T, Thulliez P, Brezin AP, Dupouy-Camet J. 2009. Contributions of immunoblotting, real-time PCR, and the Goldmann-Witmer coefficient to diagnosis of atypical toxoplasmic retinochoroiditis. J Clin Microbiol 47:2131–2135. doi: 10.1128/JCM.00128-09. [DOI] [PMC free article] [PubMed] [Google Scholar]