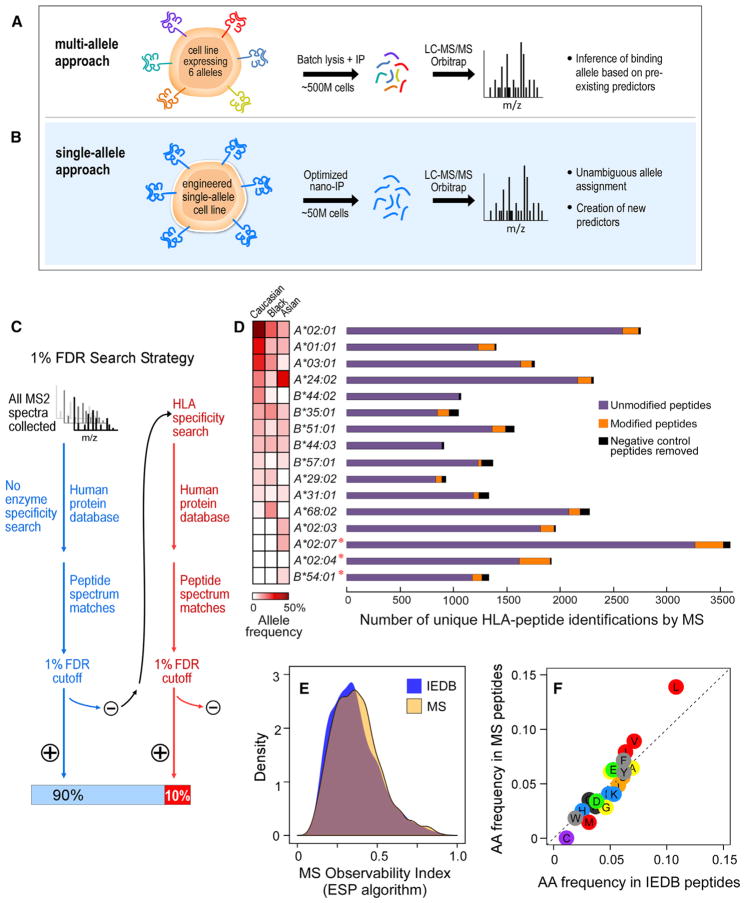
Figure 1. An Efficient Sample-Processing and -Analysis Pipeline for HLA Peptide Sequencing.
(A) Overview of the standard multi-allele workflow. Cells (~500 million [M]) expressing multiple class I HLA alleles are lysed, and HLA-associated peptides are immunopurified with a pan-anti-HLA antibody. The complex mixture of HLA peptides is sequenced via LC-MS/MS, and the allele-binding assignments are inferred from previous knowledge.
(B) In our single-allele approach, B721.221 cells (~50 M), are transduced to express only one HLA allele. Immunopurified peptides are analyzed by LC-MS/MS and sequenced via an HLA-allele-specific database search.
(C) Schema of the HLA-specific database search strategy.
(D) HLA-class-I-associated peptide identifications from 16 single-HLA-expressing cell lines. Total numbers of unmodified (purple), modified (orange), and negative control (black) peptides identified per allele are shown. Allele frequencies among Caucasian, Asian, and Black populations are shown. An asterisk denotes alleles for which LC-MS/MS experiments have generated a greater number of peptides than what is reported in the Immune Epitope Database.
(E) To evaluate LC-MS/MS bias, we calculated the “MS observability index,” as measured by the ESP algorithm (Fusaro et al., 2009), for IEDB (blue) and MS (orange) peptide datasets. Distributions of the MS observability are displayed.
(F) Amino acid frequencies within peptides reported in our single-allele dataset are compared to amino acid frequencies in peptides reported in IEDB. See also Figure S1.