Figure 1.
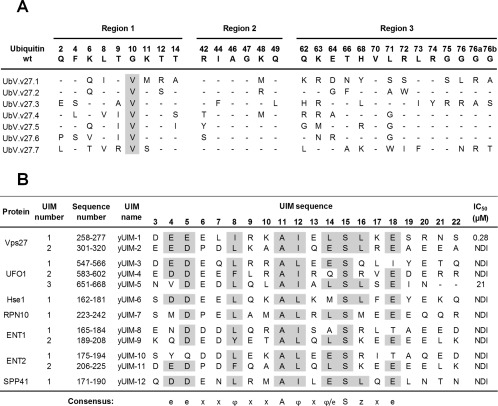
Selective binding of UbV.v27.1 to yUIM‐1. (A) Sequence alignment of UbV.v27.1 and other UbVs selected for binding to yUIM‐1. The alignment shows only those positions that were diversified in the UbV library, and positions that were conserved as the wt sequence are shown as dashes. Sequences showing conservation across selected UbVs are highlighted in grey. (B) Sequence alignment of yeast UIMs and IC50 values for inhibition of UbV.v27.1 binding to immobilized yUIM‐1. Residues (arbitrarily numbered from 3 to 22) that conform to the UIM consensus are highlighted in grey, and the consensus is shown below (e is an acidic residue, φ is a hydrophobic residue, z is a bulky hydrophobic or polar residue with high aliphatic content, A is alanine, S is serine, and x is a helix‐favoring residue). “Sequence number” denotes the position of each UIM domain in the full‐length protein sequence according to the UniProt database.14 IC50 values were defined as the concentration of solution‐phase GST‐UIM fusion protein that inhibited 50% of the UbV.v27.1 binding to immobilized yUIM‐1. “NDI” denotes “no detectable inhibition” with 60 µM GST‐UIM.
