Figure 7. GSK3β-Mediated Phosphorylation of CLASP2 Affects Dab1 Binding.
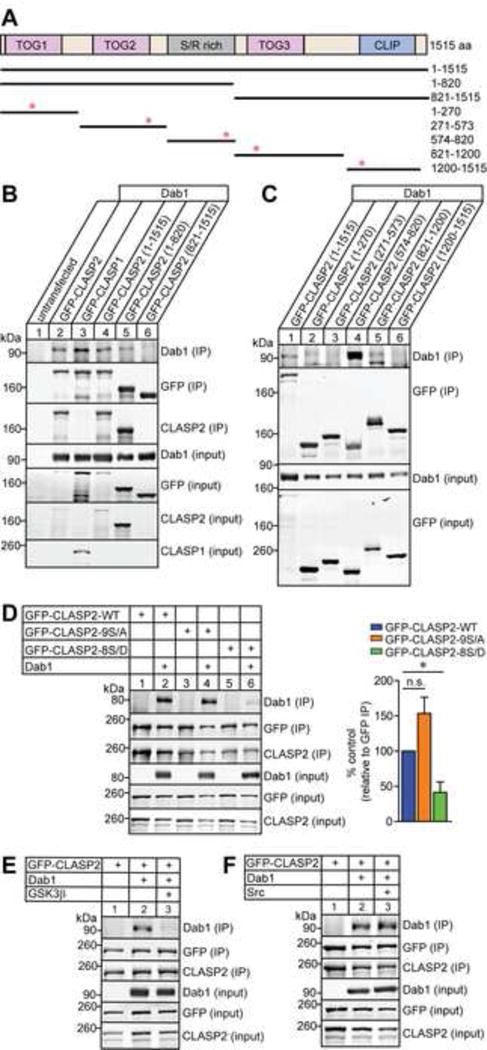
(A) Schematic of human CLASP2α domain organization and GFP-tagged CLASP2 constructs used for immunoprecipitation with corresponding amino acid (aa) residues on the right. Red asterisks indicate the position of the putative NPxY motifs in CLASP2.
(B–C) HEK293T cells were co-transfected with Dab1 and GFP-CLASP2 plasmids. Cell lysates were collected 48 h post-transfection and immunoprecipitated with GFP antibody and immunoblotted for Dab1, GFP and CLASP2. Both CLASP1 (lane 3) and CLASP2 (lane 2 and 4) bind to Dab1 and the binding region for Dab1 is within the first 820 amino acids (lane 5) of CLASP2 (B). CLASP2 specifically binds to Dab1 at the S/R domain (lane 4) between 574-820 amino acids (C).
(D) HEK293T cells were co-transfected with Dab1 and GFP-CLASP2 wild type (WT) or GFP-CLASP2-9S/A or -8S/D phospho-mutants. Cell lysates were collected 48 h post-transfection and immunoprecipitated with GFP antibody and immunoblotted for Dab1, GFP and CLASP2. The phospho-mimetic CLASP2-8S/D mutant (lane 6) showed less Dab1 binding compared to CLASP2-WT (lane 2) or the phospho-resistant CLASP2-9S/A (lane 4) mutant. These results were quantified as relative to the immunoprecipitated level of GFP (n = 3 independent experiments).
(E) The addition of constitutively active GSK3β (lane 3) abolished CLASP2 binding to Dab1 (n = 2 independent experiments).
(F) Phosphorylation of Dab1 with constitutively active Src (lane 3) did not affect CLASP2 binding to Dab1 (n = 3 independent experiments).
Data are means ± SEM and statistical significance was assessed using one-way ANOVA (*p < 0.05). n.s. represents not significant.
