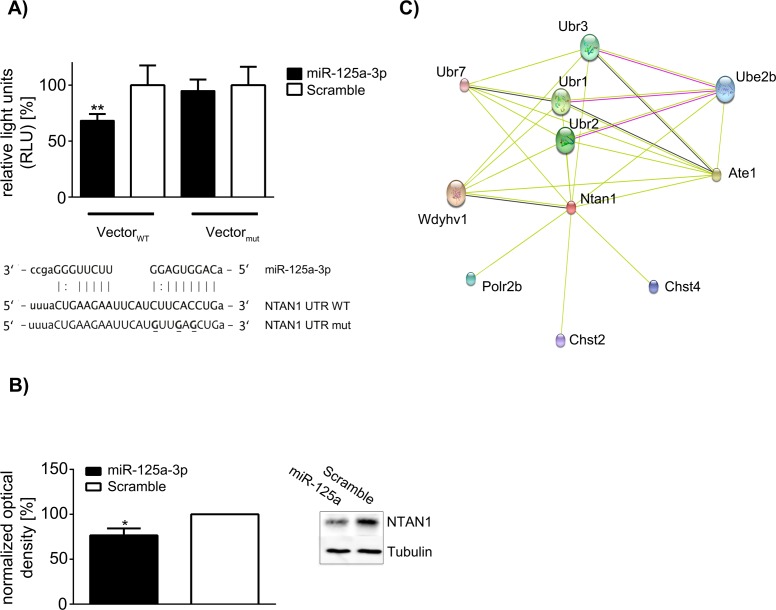
Fig 2. microRNA-125a-3p targets NTAN1 and regulates it on protein level.
microRNA-125a-3p shows partial homology to the 3´UTR of Ntan1 mRNA. To destroy the sequence compatibility in the seed region, 3 bases were exchanged (bold, underlined). MEF cells were co-transfected with the microRNA-125a-3p or a scramble sequence and the psiCheck2 vector carrying either the wild type Ntan1 3´UTR or the mutated sequence. Relative luminescence units (RLU) were determined after 72 h. Renilla luciferase signal was normalized against firefly luciferase. Relative luminescence units were calculated as a percentage of signal in scramble transfected cells (A). Raw264.7 cells were transfected with microRNA-125a-3p or a scramble sequence (5 nM), and NTAN1 protein levels were determined by western blot after 72 h. Densitometric analysis was performed with normalization against Tubulin. One representative blot is shown (B). In an interaction network, which was built from the STRING database, NTAN1 is shown to be associated with the N-end rule pathway. The observed interactions are based on experimental data (purple edges), co-expression (black edges) and textmining (green edges). Only direct interactions with an interaction score of minimum 0.4 (i.e. medium confidence) are shown. Interaction partners include arginyltransferase 1 (Ate1) and members of the ubiquitin protein ligase E3 component n-recognin (Ubr) family (C). Data are shown as mean & SD of at least four independent experiments. Statistical tests were one-way ANOVA with post-hoc intergroup comparison (A) and Mann Whitney U test (B). **p<0.01 Vector WT + miR-125a-3p vs. Vector Mutant + miR-125a-3p (A), *p<0.05 miR-125a-3p vs. Scramble (B).