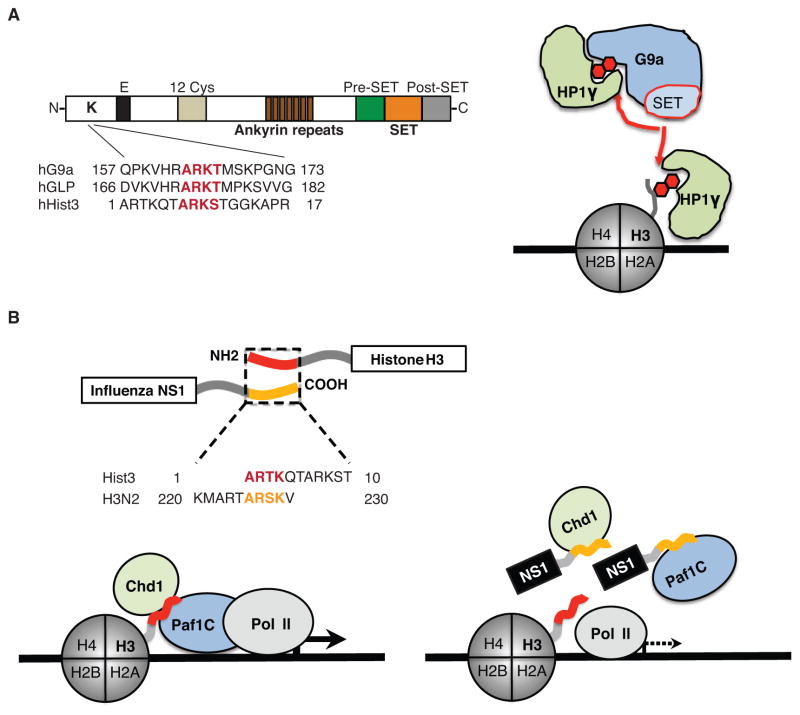
Figure 1. The structure and function of histone mimics.
A. The methyltransferases G9a and GLP possess a functional histone mimic. The histone-like sequence (red letters) is localized within the N-terminal domain of the proteins (Sampath et al. 2007). The G9a histone mimic methylation (red hexagon) is mediated in cis by the catalytic SET domain (Sampath et al. 2007) that is flanked by pre- and post-SET domains. The ankyrin repeat domain is involved in G9a interaction with methylated histones (Collins et al. 2008), and the methylated histone mimic in G9a (red hexagon) binds to the chromodomain-containing protein HP1γ that can also interact with methylated histone H3 (red hexagon). B. The NS1 proteins of the influenza A H3N2 virus possess a functional histone H3K4-like sequence. The histone-like sequence of NS1 (yellow letters) is localized within the non-structured carboxy-terminus of the protein whereas the homologous H3 sequence (red letters) is localized within the amino-terminus of histone H3 (Marazzi et al. 2012). The NS1 histone mimic (yellow tail) is present in the nucleus (Greenspan et al. 1988) where it interacts with Paf1 and Chd1 proteins. Interaction with Chd1 depends on NS1 lysine methylation, whereas Paf1 can bind to the unmethylated or methylated NS1 histone mimic. The pattern of Paf1 and Chd1 binding to NS1 is similar to these protein interactions with histone H3. The schematic model describes a putative mechanism of NS1 interference with Paf1-mediated transcription of virus-induced genes.