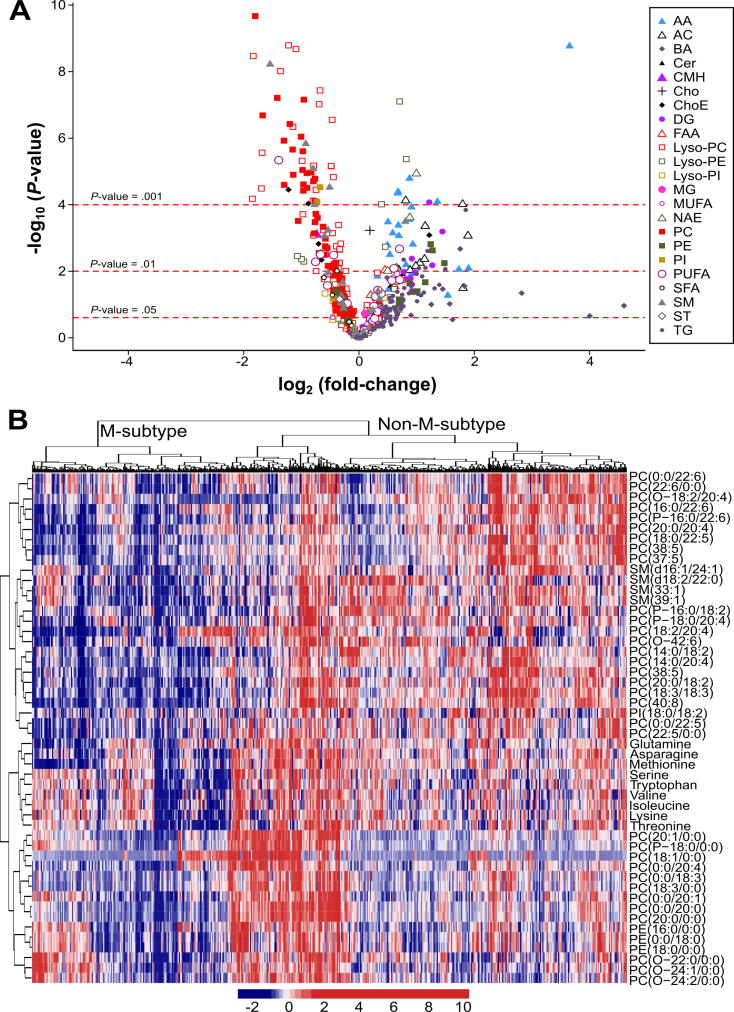
Figure 4. Identification of a subset of NAFLD patients showing a Mat1a-KO serum metabolomic profile.
A) Volcano plot representation indicating the -log10(P-value) and log2(fold-change) of individual serum metabolic ion features of MAT1A-KO compared to WT mice. AA, amino acids; AC, acyl carnitines; BA, bile acids; Cer, ceramides; CMH, monohexosylceramides; Cho, cholesterol; ChoE, cholesteryl esters; DG, diglycerides; FAA, fatty acyl amides; PC, phosphatidylcholines; Lyso-PC, lyso-phosphatidylcholines; PE, phosphatidylethanolamines; Lyso-PE, lyso-phosphatidylethanolamines; PI, phosphatidylinositols; Lyso-PI, lyso-phosphatidylinositols; MG, monoglycerides; SFA, MUFA and PUFA, saturated, monounsaturated and polyunsaturated fatty acids, respectively; NAE, N-acylethanolamines; SM, sphingomyelins; ST, steroids; TG, triglycerides. B) Heatmap representation of the serum metabolomic profile from 535 patients with biopsy-confirmed NAFLD. Each data point corresponds to the relative ion abundance of a given metabolite (vertical axis) in an individual patient’s serum. Metabolite selection is based on the top 50 serum metabolites that more significantly differentiated between MAT1A-KO and WT mice. The hierarchical clustering is based on optimum average silhouette width, obtaining the classification of the samples into two groups: first cluster resembles the serum metabolomic profile observed in the MAT1A-KO mice (M-subtype), while second cluster shows a different metabolomic profile (non-M-subtype).