Figure 5.
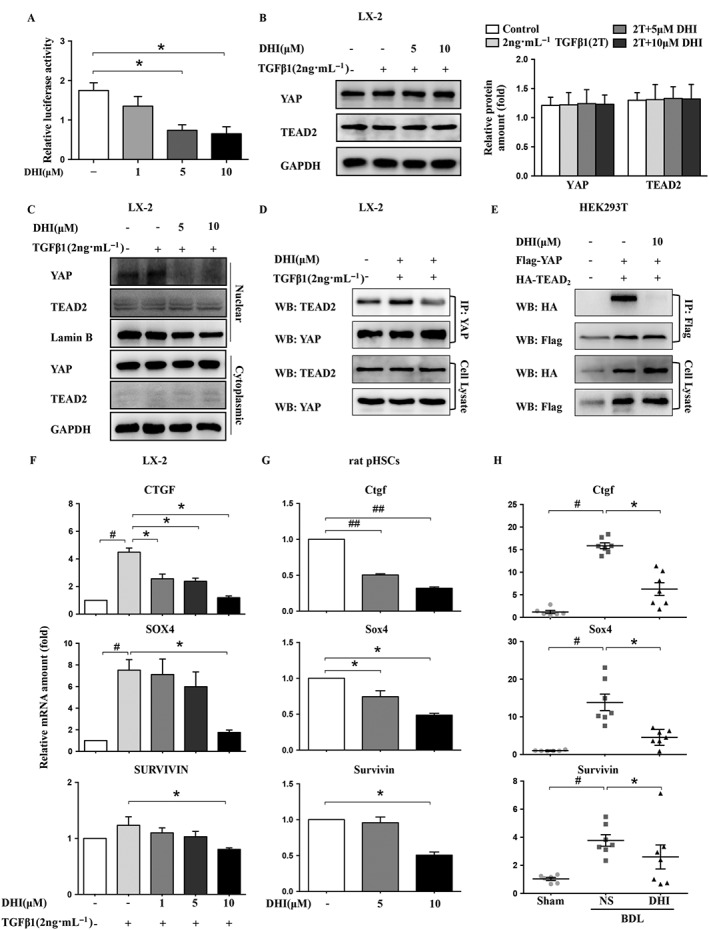
DHI interrupted the YAP/TEAD2 complex and suppressed the expression of its downstream fibrogenic genes in HSCs. LX‐2 cells were co‐transfected with pcDNA3.1‐YAP, pcDNA3.1‐TEAD2 and pGL4.17‐CTGF (A) with Renilla, and then, the luciferase activity was detected using a dual‐luciferase reporter assay, and normalized against the activity of Renilla. Western blot analysis of YAP and TEAD2 expression in LX‐2 cells (B). GAPDH served as a loading control. (C) The nuclear and cytoplasmic proteins of LX‐2 cells were separated after DHI treatment and analysed by western blotting. (D) The interaction between YAP and TEAD2 was investigated using immunoprecipitation assays of LX‐2 cells. (E) YAP‐Flag was co‐transfected with TEAD2‐HA into HEK293T cells. Whole cell extracts were immunoprecipitated with anti‐Flag and blotted with an anti‐HA antibody. The mRNA expressions of the YAP downstream genes CTGF/Ctgf, survivin and SOX4/Sox4 in LX‐2 cells (F), rat pHSCs (G) and rat liver samples (H) were detected by real‐time PCR assays and normalized against GAPDH/Gapdh. The in vitro data are expressed as the mean ± SEM of five independent assays, #P < 0.05; significantly different from the control group and *P < 0.05; significantly different from the TGFβ1 treatment group in LX‐2 cells; *P < 0.05; significantly different from the control group in rat pHSCs; the in vivo values are expressed as the mean ± SD (n = 6 in sham group; n = 7 in BDL‐NS/BDL‐DHI group), #P < 0.05; significantly different from sham group, *P < 0.05; significantly different from BDL‐NS group; ANOVA followed by Tukey's test.
