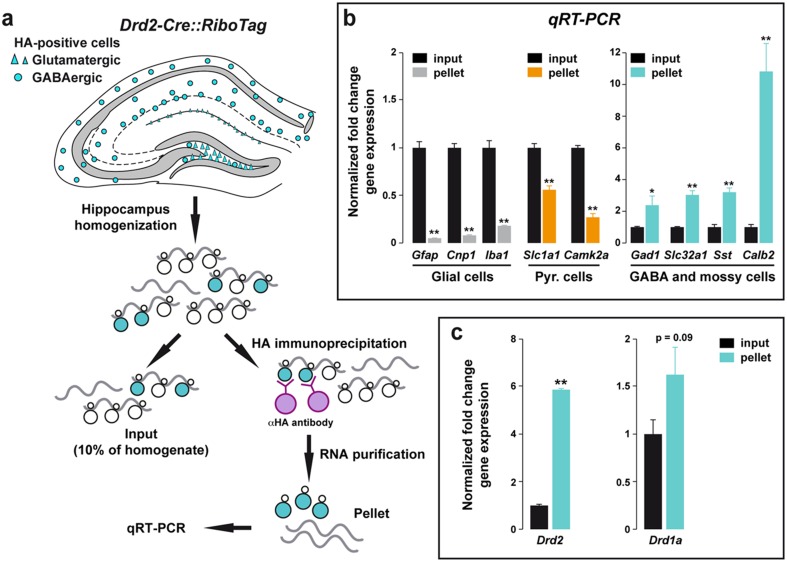
Fig. 6.
D2R-expressing cells are enriched in Drd1 mRNA. a D2R-expressing cells, either glutamatergic (triangles) or GABAergic (circles), contain ribosomes tagged with the HA epitope in Drd2-Cre::RiboTag mice. After hippocampus homogenization, 10 % of the lysate was saved as input fraction (containing all mRNAs), while the mRNAs bound to tagged-ribosomes were isolated through HA-immunoprecipitation (pellet fraction). b Quantitative RT-PCR analysis of mRNAs isolated following HA immunoprecipitation from hippocampi of Drd2-Cre::RiboTag mice. All genes were normalized to β-actin. Data are expressed as the fold change comparing the pellet fraction versus the input. Negative control genes, including glial markers (Gfap, Cnp and Iba1; grey bars) and pyramidal cell markers (Slc1a1 and Camk2a; orange bars) were de-enriched in the pellet samples, whereas the positive control genes including GABAergic and mossy cell markers (Gad1, Slc32a1, Sst and Calb2; cyan bars) were enriched in the pellet compared to the input fraction. c Quantitative RT-PCR analysis of Drd2 and Drd1 genes after HA immunoprecipitation from hippocampi of Drd2-Cre::RiboTag mice (n = 6 mice). Data are analyzed by two tailed Student t test. *p < 0.05, **p < 0.001 pellet vs. input