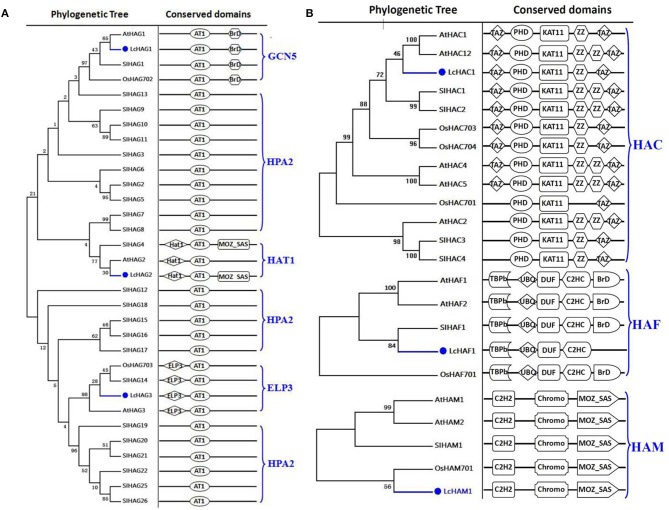
Figure 1.
Maximum likelihood phylogenetic trees and schematic diagrams for domain composition of HAT proteins predicted from Litchi chinensis (Lc), Arabidopsis thaliana (At), Oryza sativa (Os), and Solanum lycopersicon (Sl). (A) Phylogenetic tree and schematic diagrams for domain composition of HAG group. AT1 (PF00583) and C-terminal BrD (PF00439) are conserved domains of GCN5-like members; N-terminal ELP (IPR006638), and C-terminal AT1 are domains of ELP3-like; N-terminal Hat1_N (PF10394) and C-terminal AT1 are motifs of HAT1-like members while the only AT1 domain is of HPA2-like proteins. (B) Phylogenetic trees and schematic diagrams for domain composition of HAC, HAF, and HAM groups. KAT11 (PF08214), PHD-finger (PF00628), and TAZ (PF02135) are conserved domains of HAC proteins. N-terminal kinase (PF09247) (TBPb), ubiquitin, UBQ (PF00240), zinc-finger C2HC (PF01530), and C-terminal bromo BrD (PF00439) are conserved domains of HAFs. N-terminal Chromo (PF00385), C2H2 (PF00096), and C-terminal MOZ_SAS (PF01853) domains are typical of HAMs. The phylogenetic tree was constructed based on the amino acids sequences with 1,000 bootstrapping replicates.