Fig. 6. Fusion of complementary genetic circuits.
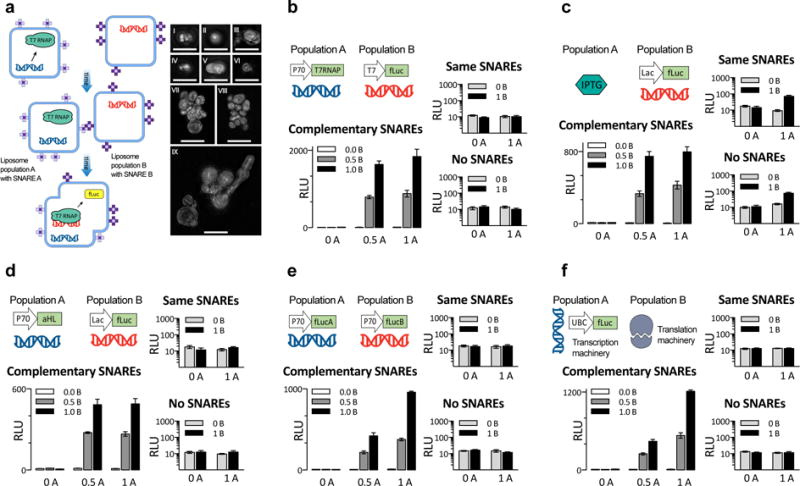
a. General scheme for SNARE-mediated liposome fusion. We created two populations of liposomes, A and B, decorated with complementary SNARE protein mimics in their outer leaflet. The images to the right, in sub-panels I through IX, are maximum-intensity projections of structured illumination microscopy (SIM) z-stacks of liposomes membrane-labeled with rhodamine, bearing complementary SNARE pairs, and fused for 4 hours. All images from panels I through IX represent separate fields of view. Scale bars, 5 μm. All liposomes in this figure, except f, contained bacterial TX/TL components. b–f. Five different instantiations of the liposome fusion concept, exploring several ways to distribute genetic circuits across fusable liposomes, with two different populations of liposomes at three occupancy levels for each case. b. Mixing of constitutively expressed T7 RNA polymerase with firefly Luciferase under T7 promoter. c. Mixing of a non-membrane-permeable small molecule activator IPTG with its inducible promoter, driving fLuc production. d. Mixing of a constitutively expressed membrane channel with an inducible promoter driving fLuc production, in the background of the small molecule that induces the promoter (IPTG). e. Mixing liposomes with genes encoding split protein. f. Mixing liposomes containing mammalian transcription (HeLa) and translation (HeLa) system, producing fLuc. For all five systems in b–f., experiments on the large graph are with one of a matching pair of SNARE on each population, the top of the two small panels is both liposomes with the same SNARE, and the bottom one neither population had any SNAREs. In both small graphs of b–f, the y-axis is in logarithmic scale to show the near-zero values for non-fusing liposomes. Switching which liposome contained which SNARE had no effect on the results (Fig. S25), whereas the absence of SNARE proteins or the presence of identical SNAREs on both populations hindered fusions (small graphs on b–f). Error bars indicate S. E. M. n=4 replicates.
