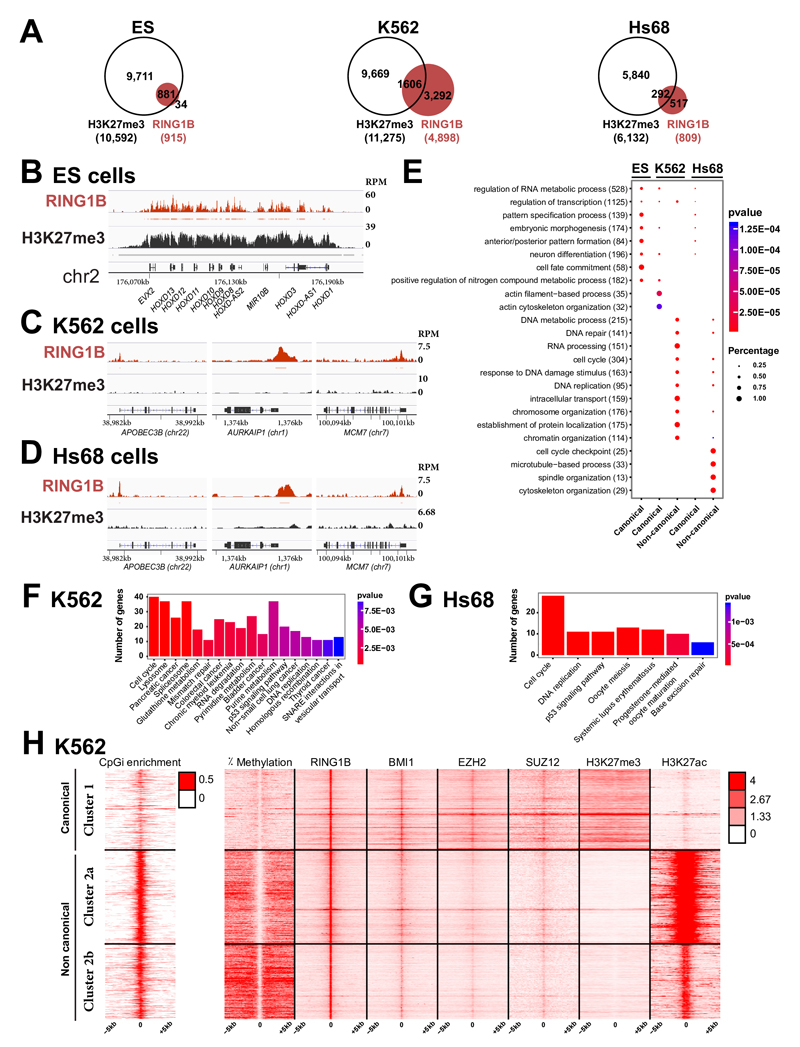
Fig. 4. PcG proteins localize to a large set of DNA-unmethylated CpG islands in the absence of H3K27me3 in differentiated human cells.
A- Venn diagram showing the overlap of RING1B target genes (in red) and genes carrying H3K27me3 (in white) in human ES cells (ES), K562 myelogenous leukemia cells and foreskin fibroblasts (Hs68). B- ChIP-Seq profiles for RING1B and H3K27me3 in ES at the HOXD complex. Significantly enriched regions are shown as horizontal bars under each ChIP-Seq track C- ChIP-Seq profiles for RING1B and H3K27me3 at a set of PRC1 loci in K562 cells. D- ChIP-Seq profiles for RING1B and H3K27me3 at a set of PRC1 loci in Hs68 cells. E- Comparative Gene Ontology (GO) analysis of canonical versus non-canonical PcG targets in ES, K562 and Hs68 cells. The total number of PcG target genes in each GO category is indicated in brackets. The fraction of bound genes and the p-values are indicated by the size and the color of each spot, respectively. F- KEGG pathways enriched in the non-canonical targets in K562 cells. G- KEGG pathways enriched in the non-canonical targets in Hs68 cells. H- Heat maps and profiles of percentage of DNA methylation, occupancies of RING1B, BMI1, EZH2, SUZ12, H3K27me3 and H3K27Ac within -/+ 5kb from RING1B peaks in K562 cells.