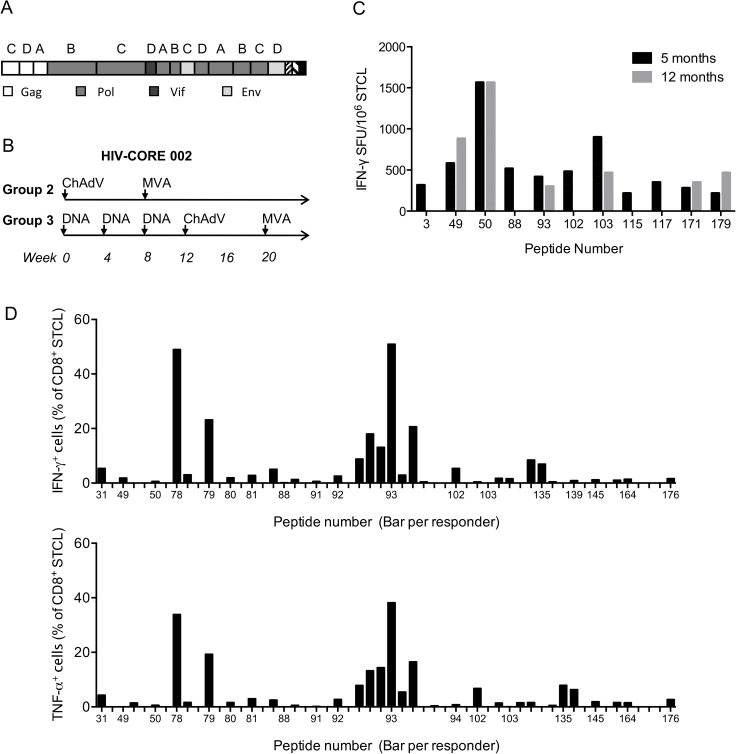
Fig 1. Conserved-region vaccine immunogen and the HIV-CORE 002 trial regimens.
(A) The box.A schematic representation of the chimaeric design of the first generation conserved immunogen HIVconsv. Capital letters above HIVconsv regions indicate the clade of origin, from which the consensus amino sequence for that region was derived. Individual HIV-1 proteins of region origin are colour coded. (B) The two vaccine regimens of the HIV-CORE 002 trial [18] that the volunteers analyzed in this study received. ChAdV–recombinant non-replicating simian (chimpanzee) adenovirus 63 ChAdV63.HIVconsv; MVA—recombinant non-replicating poxvirus modified vaccinia virus Ankara MVA.HIVconsv; and DNA–‘naked’ plasmid pSG2.HIVconsv DNA. (C and D) Optimal epitope mapping was performed using thawed vaccine-recipients’ PBMCs, which were in vitro expanded with the parental 15-mer peptides for 10 days to establish STCL. (C) Frequencies of vaccine-elicited, HIV-1-specific, in vitro 15-mer peptide (x-axis)-expanded sequential PBMC samples of several volunteers from 5 (black) and 12 (grey) months after the last vaccine administration. Specific cells were enumerated in an IFN-γ ELISPOT assay. Panel (D) overviews the frequencies of peptide-specific CD8+ STCLs over the volunteers and peptides used. Frequencies of specific cells from all tested volunteers expanded by the same 15-mer peptide are shown next to each other above each peptide given on the x-axis. Top and bottom graphs show frequencies of CD8+ T cells producing IFN-γ and TNF-α, respectively. S2 Table for the list of peptides and responding volunteers.