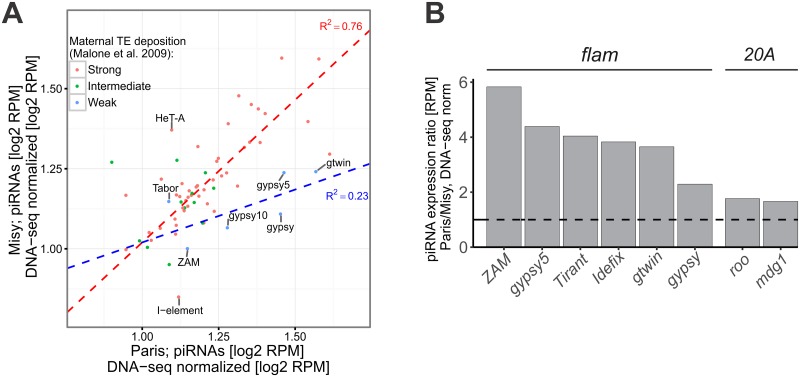
Fig 4. Uni-strand piRNA clusters produce more piRNAs in the Paris than in the Misy strain.
(A) Scatter plot of log2-transformed and RPM-normalized small RNA expression in ovaries of Paris and Misy strains. piRNA expression was additionally normalized to the log2-transformed and RPM-normalized genomic abundance of the corresponding TEs estimated as number of DNA-seq reads mapped to the canonical TE sequences. The color of dots indicates the type of TEs according to its capacity for maternal deposition in embryos according to [24]. Dashed lines depict the results of linear regression analysis for TEs with high maternal deposition (genuine germinal TEs, red) and for TEs with weak maternal deposition (genuine somatic TEs, blue). R2—adjusted squared R (P-value < 0.1). (B) Relative abundance (Paris/Misy) of a normalized number of single-mapped small RNA reads (RPM, 24–29 nt reads were considered) mapping to TE copies located within uni-strand piRNA clusters #8 (flamenco, expressed in follicular cells) and #2 (20A, expressed in the germline).