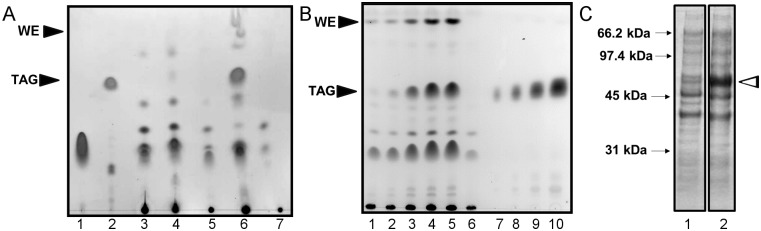
Fig 2. Detection of TAG production in the E. coli C41 (DE3) strain carrying the enzyme tDGAT and correlation with its expression.
(A) TLC showing the analysis of the lipid fractions extracted from cultures of E. coli C41 (DE3) with the plasmid constructions pET29c::Ma1 (lane 3), pET29c::Ma2 (lane 4), pET29c::Ab (lane 5), pET29c::tDGAT (lane 6) and the naked plasmid pET29c (lane 7). Oleic acid (lane 1) and trioleoylglycerol (lane 2) were loaded as control standards. Positions of TAGs and WEs are marked by black arrows. (B) TLC plate showing TAG production in the engineered strain E. coli C41 (pET29c::tDGAT) along time after IPTG induction. Lipid extractions from 6.5 mg of dry biomass collected 0, 30, 60, 90 and 120 minutes after induction with 100 mM IPTG at OD600nm = 0.5 were loaded in lanes 1–5, respectively. A similar lipid extraction from wild type E. coli was loaded in lane 6. Different amounts of purified commercial TAGs (4.5; 18; 36 and 72 μg) were loaded in lanes 7–10 in order to estimate the TAG production by densitometric assay. The black arrows point to the migration distance of the TAGs and WEs. (C) SDS-PAGE electrophoretic gel of whole cell lysates of E. coli (pET29c::tDGAT) collected after induction periods of 0 and 90 minutes (lanes 1 and 2). Molecular masses (in kilodaltons) are indicated at the left. The band indicated by the white arrowhead corresponds to the protein tDGAT.