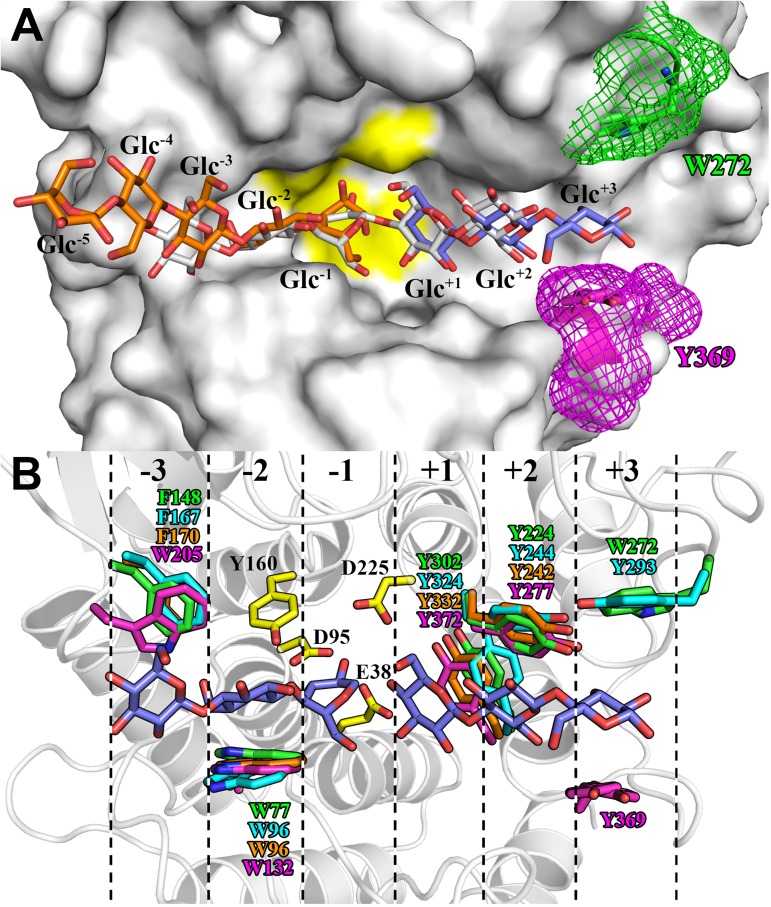
Fig 6. AfmE1 substrate-binding cleft.
(A) Molecular surface of AfmE1 with its stacking residue Trp272 in subsite +3 represented as sticks with carbon atoms in green. The stacking residue Tyr369 in the corresponding region of CelA from C. thermocellum is similarly represented in magenta to evidence the differences in the subsite +3 configuration of these enzymes. The region containing the catalytic residues is highlighted in yellow. The substrate molecules, represented as deduced from the complex of CelA with cellopentaose (white) and cellotriose (blue) (PDB code 1KWF), as well as of BcsZ with cellopentaose (orange) (PDB code 3QXQ), are shown as sticks to indicate the position of the subsites. (B) AfmE1 substrate-binding cleft highlighting the catalytic (yellow) and the glucosyl-stacking residues (green) in its six subsites (dashed lines). The corresponding stacking residues of the proteins CMCax, BcsZ and CelA are shown in cyan, orange and magenta, respectively. The position of the glucosyl residues (blue) occupying the six subsites was predicted from the complex CelA-substrate [44].