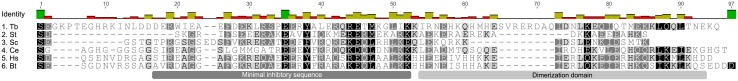
Fig 1. TbIF1 shares homology with IF1 proteins from various model organisms.
Using the alignment program MAFFT, the amino acid sequence of IF1 from T. brucei (Tb) is compared to those of Solanum tuberosum (St, [20]), Saccharomyces cerevisiase (Sc, [22]), Caenorhabditis elegans (Ce, [21]), Human sapiens (Hs, [44]) and Bos taurus (Bs, [19]) IF1s. Identical and conservatively substituted resides are shaded in black and grey, respectively. Predicted mt import signal sequences were removed from each coding sequence prior to the analysis. The minimal inhibitory sequence [42] and dimerization domain of bovine IF1[45] are indicated. The green, yellow, and red bars above the alignment indicate when the nucleotides at each position have identities of 100%, between 100% and 30%, and <30%, respectively.