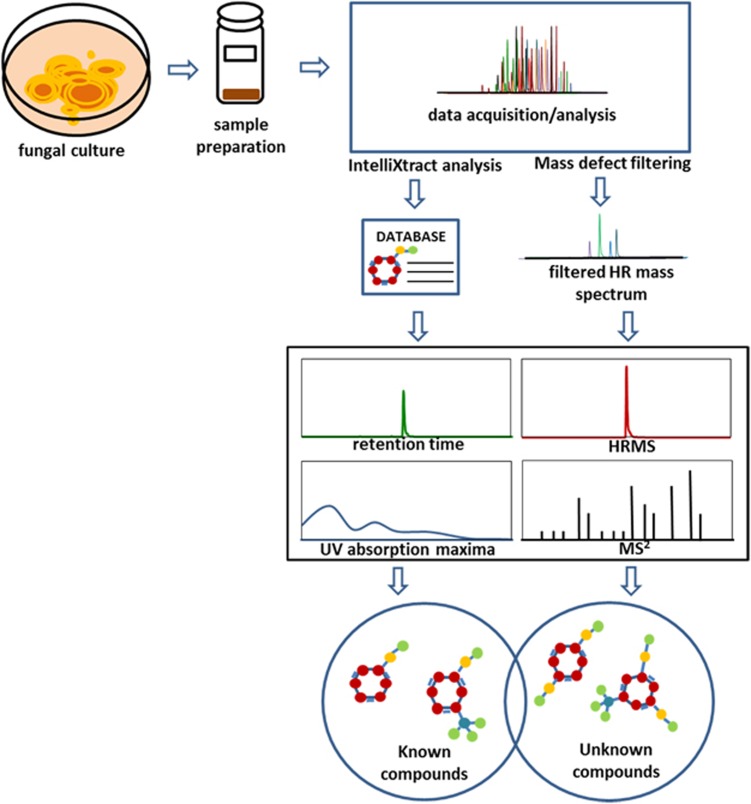
Scheme 1.
Proposed workflow for dereplication of fungal extracts including a 5 min ultraperformance liquid chromatography-photodiode array-high resolution tandem mass spectrometry (UPLC-PDA-HRMS-MS/MS) methodology. HRMS and MS/MS (collision-induced dissociation (CID) 30) data were acquired by LTQ Orbitrap using data-dependent scan. A postacquisition data analysis using IntelliXtract, an add-in feature of ACD MS Manager, cross-references the molecular ion peaks and retention times to identify compounds known to the in-house database. For targeted analysis of a specific class of compounds, a user-defined mass defect filtering criterion was designed based on accurate masses and mass defects centered around a core structure. The HRMS data were filtered with Compound Discoverer software to identify components related to the known compounds. The identity of the known compounds were verified based on HRMS, MS/MS fragmentation pattern, retention time and UV absorption maxima. The tentative identity of the discovered unknown compounds were assigned based on the interpretation of their CID-MS/MS (30 eV) fragmentation patterns.