Abstract
As glutathione (GSH) plays an essential role in growth and symbiotic capacity of rhizobia, a glutathione synthetase (gshB) mutant of Rhizobium leguminosarum biovar viciae 3841 (Rlv3841) was characterised. It fails to efficiently utilise various compounds as a sole carbon source, including glucose, succinate, glutamine and histidine, and shows 60%–69% reduction in uptake rates of glucose, succinate and the non-metabolisable substrate α-amino isobutyric acid. The defect in glucose uptake can be overcome by addition of exogenous GSH, indicating GSH, but not its bacterial synthesis, is required for efficient transport. GSH is not involved in the regulation of the activity of Rlv3841's transporters via the global regulator of transport, PtsNTR. Although lack of GSH reduces transcription of the branched amino acid transporter, this was not the case for all uptake transport systems, for example, the amino acid permease. This suggests GSH alters activity and/or assembly of transport systems by an unknown mechanism. In interaction with plants, the gshB mutant is not only severely impaired in rhizosphere colonisation, but also shows a 50% reduction in dry weight of plants and nitrogen-fixation ability. This reveals that changes in GSH metabolism affect the bacterial–plant interactions required for symbiosis.
Keywords: Rhizobium, glutathione, nitrogen fixation, pea, nodulation
Glutathione is needed for bacteria to fix nitrogen efficiently in symbiosis with legumes.
INTRODUCTION
Rhizobia are gram-negative bacteria that are able to establish an effective nitrogen-fixing symbiosis with their plant hosts (Oldroyd et al.2011; Udvardi and Poole 2013). A host plant produces a specific chemical signal molecule (flavonoid) that is perceived by the bacterium, and in response rhizobia produce lipo-chitooligosaccharides (Nod factors) leading to root hair curling which surrounds the bacteria (Long 1996). Following endocytosis into an infected cortical cell, rhizobia are engulfed by a plant-derived membrane (symbiosome membrane), to form a symbiosome (Clarke et al.2015). Inside symbiosomes, rhizobia differentiate into nitrogen-fixing bacteroids. During the infection process and nodule formation, rhizobia are greatly dependent on glutathione (GSH) to protect themselves against oxidant damage (Harrison et al.2005). The thiol tripeptide GSH (L-glutamyl-L-cysteinylglycine) is abundant in living cells where it acts as an antioxidant either directly, by interacting with reactive oxygen and nitrogen species (ROS and RNS, respectively) and electrophiles, or by operating as a cofactor for various enzymes (Ribeiro et al.2015). As well as being synthesised within the bacteria, GSH is produced by plant roots during the process of nodule formation (Becana et al.2010; Matamoros et al.2015). GSH biosynthesis in bacteria is a two-step process catalysed by ATP-dependent enzymes. Glutamic acid and cysteine are conjugated by γ-glutamylcysteine synthetase (encoded by gshA), and its product, γ-glutamylcysteine, is linked to glycine to form GSH in a reaction catalysed by glutathione synthetase (encoded by gshB).
In addition to its antioxidant role in nodule formation, it has been shown that free-living rhizobia, such as Rhizobium etli, R. tropici, Ensifer (Sinorhizobium) meliloti and Bradyrizobium sp. require GSH to combat environmental stress and to efficiently utilise various compounds, such as glucose, succinate, glutamine, alanine and histidine, as their sole carbon source (Riccillo et al.2000; Muglia et al.2008). In R. etli, GSH is required for glutamine utilisation and symbiotic effectiveness (Taté et al.2012).
Rhizobia are known to be especially rich in ATP-binding cassette (ABC) transporters (Walshaw and Poole 1996). In R. leguminosarum biovar viciae 3841 (Rlv3841), amino acid transport occurs predominantly via two ABC transport systems, termed the general amino acid permease (Aap) and the branched-chain amino acid permease (Bra) (Hosie et al.2002). Glucose is transported into Rlv3841 cells by at least three different ABC transport systems (Untiet et al.2013). In R. leguminosarum, transport of L-malate, fumarate and succinate occurs via the C4-dicarboxylic transport (Dct) system (Reid and Poole 1998). It is worth noting that PtsNTR acts as a global regulator of many functions in the cell, including regulating ABC transport systems at the post-transcriptional level. The first component of this system, PtsP, is phosphorylated by PEP and transfers this phosphate to PtsN1 and PtsN2 via a small protein Npr/Hpr. Inactivation of PtsP, Npr or both copies of PtsN results in a severe inhibition of transport of amino acids by Aap and Bra (Prell et al.2012; Untiet et al.2013). The precise mechanism by which PtsN regulates ABC transporters is unknown, but the overlap with the phenotype of strains altered in GSH metabolism indicated the possibility that PtsNTR might control ABC transporters via GSH levels.
Given the importance of GSH in many rhizobia during symbiosis, we consider it important to understand the effects of GSH on carbon compound transport and metabolism in both free-living and symbiotic R. leguminosarum.
MATERIALS AND METHODS
Bacterial growth and media
Strains, plasmids and primers used in this study are listed in Table 1. Rhizobium leguminosarum strains were grown at 28°C in either Tryptone Yeast extract (TY) (Beringer 1974) or acid minimal salts medium (AMS) (Poole et al.1994) with 10-mM D-glucose (Glc) or 10-mM di-sodium succinate (Suc) as carbon sources and 10-mM NH4Cl as a nitrogen source, unless otherwise described. They were grown on AMS agar containing 5-mM glutamine (Gln), glutamate (Glu), histidine (His), alanine (Ala) or asparagine (Asn) as sole carbon and nitrogen source.
Table 1.
Strains, plasmids and primers.
| Strain, plasmid or primer | Description | Reference, source or DNA sequence (5΄-3΄)a |
|---|---|---|
| Rlv3841 | R. leguminosarum biovarviciae, Strr | Johnston and Beringer (1975) |
| LMB599 | Rlv3841 gshB::pKGshB, Strr, Neor | This study |
| LMB675 | LMB599[pGshB], Strr, Neor, Tetr | This study |
| PtsP107 | Rlv3841 Tn5::ptsP; Strr, Neor | Prell et al. (2012) |
| pRK415-1 | Plasmid; IncP broad host range cloning vector, Tetr | Karunakaran et al. (2006) |
| pK19mob | Plasmid; pUC19-derivative, plasmid,lacZ mob; Kmr | Karunakaran et al. (2010) |
| pRK2013 | Helper plasmid for mobilising plasmids; Kmr | Figurski and Helinski (1979) |
| pKGshB | Plasmid; pr1373/pr1374 PCR product cloned in pK19mob, Kmr | This study |
| pGshB | Plasmid; pr1490/pr1491 PCR product cloned in pRK415-1, Tetr | This study |
| pK19A | Primer; pK19mob plasmid mapping primer | Karunakaran et al. (2010) |
| p1022, p1023 | Primers for qRT-PCR of aapJ | Mulley et al. (2011) |
| p1028, p1029 | Primers for qRT-PCR of aapQ | Mulley et al. (2011) |
| p1030, p1031 | Primers for qRT-PCR of braC | Mulley et al. (2011) |
| p1032, p1033 | Primers for qRT-PCR of braD | Mulley et al. (2011) |
| p1034, p1035 | Primers for qRT-PCR of braE | Mulley et al. (2011) |
| p1038, p1039 | Primers for qRT-PCR of braG | Mulley et al. (2011) |
| pr1373 | Sense primer for PCR of amplifying internal fragment of RL0338 (gshB) | AAATCTAGACCGGAG CGCGTCGACCTTGC |
| pr1374 | Antisense prime for PCR of amplifying internal fragment of RL0338 (gshB) | AAAAAGCTTCGGCCGCCGACATGCATGTT |
| pr1375 | Mapping PCR primer for checking gshB mutation | CGAAGGTGAGGTCCATATCA |
| pr1490 | Sense primer for PCR of complete RL0338 (gshB) gene | TTGGATCCAACAGCGGCAACCAGGGCAT |
| pr1491 | Antisense primer for PCR of complete RL0338 (gshB) gene | TTGGATCCTCAGCCGCGCTTGCGTTCGAT |
| pr1562 | Sense primer for qRT-PCR of gyrB1 | GGCATCACCAAAAGGGAAAA |
| pr1563 | Antisense primer for qRT-PCR of gyrB1 | GCGAGGAGAATTTCGGATCA |
Restriction sites in primer sequences are underlined.
To determine a strain's ability to grow on different compounds, the method described by Taté et al. (2012) was used with the media described above (TY and supplemented AMS), with and without addition of exogenous 100 μM GSH as described by Taté et al. (2012) and photographed after 3 days’ incubation at 28°C. Antibiotics were used at the following concentrations (μg mL−1): carbenicillin (Carb), 50; gentamicin (Gm), 20; kanamycin (Km), 20; neomycin (Neo), 80 (for R. leguminosarum), or 250 (for Escherichia coli); nyastatin (Ny), 50; tetracycline (Tet), 5; spectinomycin (Spec), 100; streptomycin (Str), 500. Optical density (OD600) measurements were automatically recorded every 30 min in an Eon Microplate Spectrophotometer (Bio-Tek) and used for calculation of doubling times for each strain.
Mutants and plasmids
A mutation in gshB (RL0338) of Rlv3841 was made by plasmid integration using single crossover. A 500-bp gshB fragment was PCR amplified using primers pr1373/pr1374 (Table 1). The fragment was cloned into XbaI/HindIII sites of pK19mob, resulting in plasmid pKGshB. Plasmid pKGshB was transferred from E. coli to Rlv3841 and recombined into the genomic gshB region via single crossover to give strain LMB599. Insertion into the gshB gene was confirmed by PCR using pr1375 and a pK19mob-specific primer pK19A (Karunakaran et al.2010).
To complement the gshB mutation in LMB599, primers pr1426 and pr1427 were used to amplify the complete gshB gene from Rlv3841. The PCR product was digested with BamHI and cloned into BamHI-digested pRK415-1, resulting in plasmid pGshB. Plasmid pGshB was conjugated into LMB599 using pRK2013 as a helper plasmid to provide the transfer genes, as previously described (Figurski and Helinski 1979), to make complemented strain LMB675.
Transport assays
Rhizobium leguminosarum uptake assays were performed with 25-mM (4.625 kBq of 14C) solute (Hosie et al.2002), using cultures grown in AMS with Glc or Suc to an OD600 of ∼0.35. When required, an additional 100-μM GSH was added.
RNA isolation and quantitative RT-PCR analysis
Independent 50-mL cultures of R. leguminosarum strains were grown overnight in AMS Glc in 250-mL flasks at 225 r.p.m. to log phase (OD600 0.3–0.4), in triplicate. Cultures (12 mL) were added directly to RNAlater (Ambion) (24 mL) and centrifuged at 11 000 r.p.m., 4°C in a Sorvall SS-34 rotor centrifuge. RNA was treated twice with TURBO DNase using a DNA-free kit (Ambion). cDNA was synthesised using SuperScript II RT and random hexamers (Invitrogen, Carlsbad, California USA). Quantification of cDNA was carried out using the SensiMix SYBR No-ROX kit (Bioline), and real-time amplification of the PCR products was performed using Bio-Rad CFX real-time PCR system. Primers for amplification by qRT-PCR are detailed in Table 1. PCR conditions consisted of an initial incubation step for 3 min at 95°C, followed by 35 cycles for 5 s at 95°C, 10 s at 62°C and 5 s at 72°C. The gyrb1 gene was used as a calibrator gene and results were analysed as previously described Prell et al. (2009). Statistical analysis of data sets was performed using REST (Pfaffl, Horgan and Dempfle 2002) on data from three independent biological samples (each with three technical replicates).
Dry weight analysis and acetylene reduction
Rhizobium leguminosarum strains were used to inoculate surface-sterilised pea seeds (Pisum sativum cv. Avola) at the time of sowing. For dry weight determination, plants were grown as previously described (Poole et al.1994) in a growth room (16-h light/8-h dark) in 2-L beakers filled with sterile medium-grade vermiculite, watered with nitrogen-free nutrient solution and harvested at 6 weeks. The shoot was removed from the root and dried at 70°C in a dry-heat incubator for 3 days before being weighed. Acetylene reduction was determined at pea flowering (3 weeks) as previously described (Allaway et al.2000).
Rhizosphere colonisation
Surface-sterilised pea seeds were grown as described above, but in autoclaved boiling tubes filled with 25 mL of washed fine-grade vermiculite supplemented with 10 mL of nitrogen-free rooting solution. Inoculation with the following ratios of colony forming units (cfu) of Rlv3841 and LMB599 was performed after 7 days: 1000:0, 0:1000, 1000:1000 and 1000:10000. At 7 days post-inoculation (dpi) (14 days after planting), plants were removed from the vermiculite, shoots were cut-off and 20 mL of sterile phosphate-buffered saline was added to the root of each plant and vortexed in Multi Reax system (Heidolph) for 15 min, speed 10 (Karunakaran et al.2006). Following serial dilution in sterile phosphate-buffered saline, samples were plated onto TY medium containing Str and Ny (both Rlv3841 and LMB599 will grow) or Str, Ny and Neo (only LMB599 will grow), to give the total number of each strain in the pea rhizosphere (Barr et al.2008). Agar plates were incubated at 28°C for 3 days, colonies counted and percentage of cells recovered calculated.
RESULTS
Rlv3841 gshB mutant (LMB599) is defective in growth
A mutation was made in the GSH synthetase gene (gshB) of Rlv3841 to give strain LMB599. In liquid cultures of TY or AMS with succinate or glucose as a carbon source, GSH-deficient strain LMB599 has doubling times increased by approximately 2-fold compared to the parental strain Rlv3841 (Table 2). Addition of 100-μM GSH to LMB599 cultures reduces the doubling time in glucose-grown cultures from 12.9 to 5.5 h, to give values similar to those of wild-type Rlv3841 (approx. 5.4 h) (Table 2). This shows that the effects of a mutation in GSH synthesis on bacterial growth are overcome by addition of exogenous GSH.
Table 2.
Growth of wild-type Rlv3841 and GSH-deficient mutant LMB599.
| Doubling time (h) | ||||
|---|---|---|---|---|
| Strain | TY | AMS Suc | AMS Glc | AMS Glc/GSH* |
| Rlv3841 | 4.8 ± 0.0a | 6.8 ± 0.0a | 5.3 ± 0.1a | 5.4 ± 0.0a |
| LMB599 | 8.0 ± 0.4b | 13.7 ± 0.6b | 12.9 ± 0.2b | 5.5 ± 0.1a |
Strains grown in TY or AMS minimal medium supplemented with glucose (Glc) or succinate (Suc) as the sole carbon source. *100-μM GSH added. Data are averages (±SEM) from three independent cultures. a,b indicate significant difference (P ≤ 0.01).
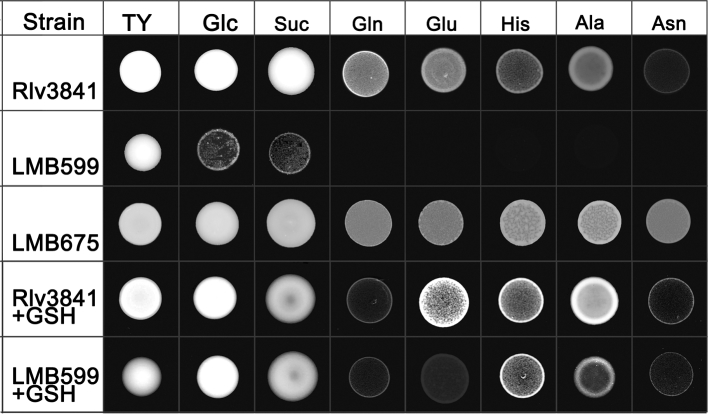
On solid media, LMB599 grows poorly on AMS with glucose and succinate as a sole carbon source, while growth is undetectable with amino acids glutamine, glutamate, histidine, alanine and asparagine as sole sources of carbon and nitrogen (Fig. 1). Growth of LMB599 on all the media assayed is restored to wild-type Rlv3841 levels by complementation with a cloned full-length copy of gshB carried on a plasmid (strain LMB675; LMB599[pGshB]) (Fig. 1). Indeed, in LMB675 growth on histidine, alanine and asparagine appears to be better than in wild-type Rlv3841. Addition of 100-μM GSH also restores the growth of LMB599 on glucose and succinate, although exogenous GSH has a slightly less clear-cut effect on amino acid transport. In the gstB mutant, slight growth is seen on histidine and alanine while there is little discernible growth on glutamine, glutamate and asparagine as sole sources of carbon and nitrogen. Addition of exogenous GSH also adversely affects the growth of Rlv3841on glutamine a sole carbon and nitrogen source (Fig. 1).
Figure 1.
Growth on different media of wild-type Rlv3841, GSH-deficient mutant LMB599 and complemented mutant LMB675. As described by Taté et al.(2012), cultures were spotted onto TY agar (complete medium) or AMS agar (minimal medium) containing glucose (Glc) or succinate (Suc) as sole carbon source; glutamine (Gln), glutamate (Glu), histidine (His), alanine (Ala) or asparagine (Asn) as sole carbon and nitrogen sources. + GSH, growth with 100-μM glutathione added exogenously. Plates were incubated at 28°C for 3 days.
GSH is required by a broad range of carbon transport systems
As LMB599 grows poorly on a range of amino acids, we tested amino acid uptake in LMB599 and compared it with that of wild-type Rlv3841. Amino acid transport was measured with the non-metabolisable amino acid α-amino isobutyric acid (AIB) because, like glutamine, it is transported exclusively by the Aap and Bra uptake systems (Hosie et al.2002). As AIB cannot be metabolised, the measurement of AIB transport is not complicated by secondary metabolic effects. Strain LMB599 shows a 69% reduction in the rate of AIB uptake (Fig. 2A).
Figure 2.
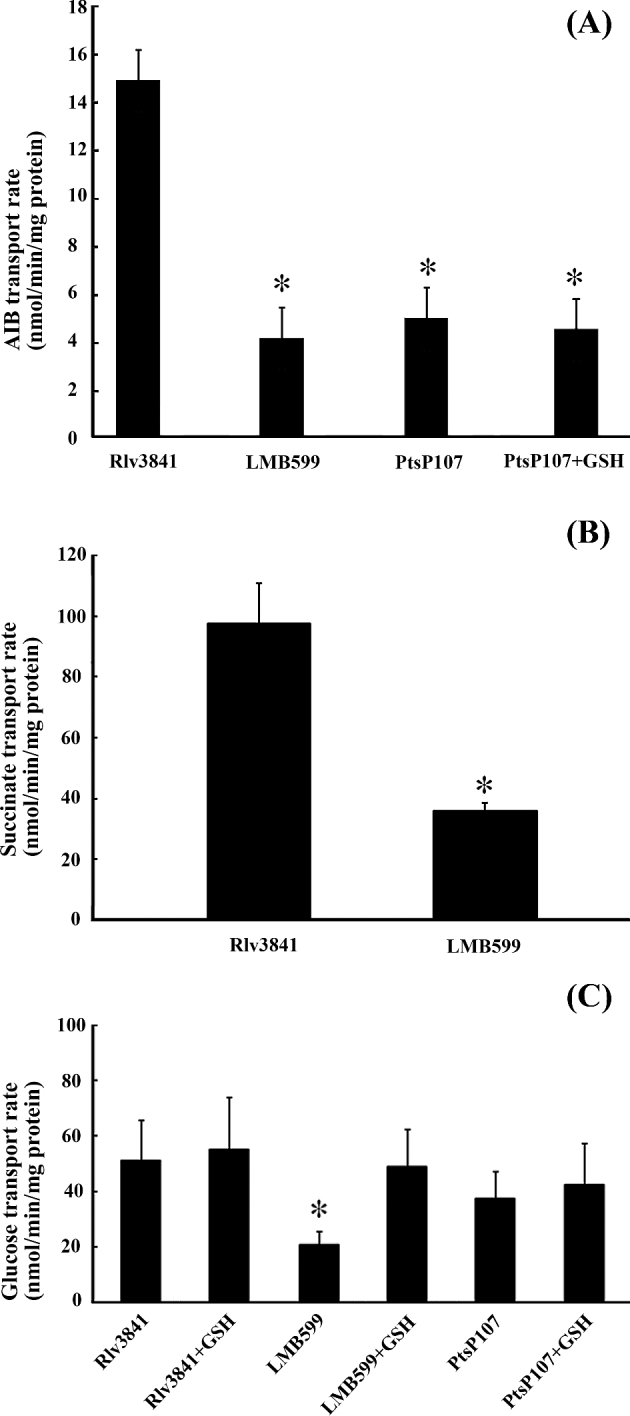
Uptake of AIB, succinate and glucose by strains of R. leguminosarum. (A) 14C-AIB uptake by wild-type Rlv3841, GSH-deficient mutant LMB599 and PtsP107 (ptsP mutant) following overnight growth in AMS Glc, with or without addition of 100-μM GSH. An asterisk indicates a significant difference compared to wild-type Rlv3841 (P ≤ 0.001). (B) 14C-Suc uptake by Rlv3841 and LMB599 following overnight growth in AMS Suc. An asterisk indicates a significant difference compared to wild-type Rlv3841 (P ≤ 0.05). (C) 14C-Glc uptake by Rlv3841, LMB599 and PtsP107 following overnight growth in AMS Glc, with or without addition of 100-μM GSH. An asterisk indicates a significant difference compared to wild-type Rlv3841 (P ≤ 0.05). All data are averages (±SEM) from ≥three independent cultures.
As growth on both glucose and succinate as a sole carbon source is also substantially reduced in strain LMB599, it may be that the effects of GSH deficiency are more general than just those observed on amino acid uptake. Uptake of succinate and glucose was measured in strain LMB599 and are reduced by 63% and 60%, respectively (Fig. 2B and C). Addition of 100-μM GSH rescues the defect in glucose transport (Fig. 2C). Thus, in comparison with Rlv3841, LMB599 shows a significant reduction in transport of AIB, succinate and glucose (Fig. 2).
Role of GSH in altering activity of ABC transport systems
PtsP has been shown to be essential for activation of a broad range of ABC transport systems (Prell et al.2012), and it is possible that this occurs via a lack of GSH. To test this hypothesis, we examined whether addition of exogenous GSH has any effect on the transport of amino acids and glucose in a ptsP mutant, strain PtsP107 (Prell et al.2012). The significantly lower rate of AIB transport in strain PtsP107 compared to Rlv3841 is not affected by the addition of 100-μM GSH (Fig. 2A). Glucose transport is not significantly different in strains PtsP107 and Rlv3841, and is unaffected by addition of exogenous GSH (Fig. 2C). GSH is found to have no role in the post-translational PTSNtr system-mediated effects on ABC transport systems.
To investigate whether transcriptional control is involved, qRT-PCR was used to determine whether the Aap and Bra transport systems, i.e. those transporting AIB, are transcriptionally affected in the GSH-deficient mutant. In LMB599, expression of the Bra transporter genes is downregulated significantly (>2-fold, P ≤ 0.05) (braC 2.97±0.55↓, braD 25.24±7.88↓, braE 3.85±1.11↓ and braG 4.95±2.41↓), while expression of genes encoding the Aap permease is not significantly (P > 0.05) slightly upregulated (aapJ 2.41±0.33↑) or not altered (<2-fold) (aapQ 1.36±0.36↑) (where ↓ shows fold decrease in relative expression and ↑ shows fold increase in relative expression compared to Rlv3841). GSH significantly downregulates the transcription of genes encoding the Bra transporter but not those of the Aap transporter which essentially show unchanged expression.
Nodulation and competitiveness in the rhizosphere of a GSH-deficient mutant
In order to assess the nodulation and nitrogen-fixing capacity of GSH-deficient mutant LMB599, Pisum sativum seedlings were inoculated with R. leguminosarum strains and the number of nodules per plant, dry weight of plants and acetylene reduction activity measured. Strain LMB599 forms a similar number of nodules as wild-type Rlv3841 (approx. 180); however, there are almost twice as many small (pink and white) nodules (approx. 75% of 177 ± 58) on LMB599-inoculated plants than for those inoculated with wild-type Rlv3841 (approx. 45% of 176 ± 23) (Table 3). The gshB mutant LMB599 shows a 50% drop in the dry weight of plants and a 50% decrease in acetylene reduction activity (Table 3). Plants nodulated with strain LMB675, in which the mutation in gshB is complemented by a full-length gshB gene cloned on a plasmid, have wild-type properties; approximately the same number of nodules per plant, same percentage of small nodules (approx. 49% of 174 ± 22) and reduce acetylene at the same rate as Rlv3841-inoculated plants (Table 3).
Table 3.
Symbiotic behaviour of wild-type Rlv3841, GSH-deficient mutant LMB599 and complemented mutant LMB675.
| Nodules | Small nodules/total | Acetylene reduction | Dry weight | |
|---|---|---|---|---|
| Strain | per plant | nodules (%)* | (μmoles acetylene per plant per h) | per plant (g) |
| Rlv3841 | 176 ± 23a | 45 ± 5a | 4.10 ± 0.10a | 2.38 ± 0.31a |
| LMB599 | 177 ± 58a | 78 ± 6b | 2.01 ± 0.29b | 1.15 ± 0.27b |
| LMB675 | 174 ± 22a | 49 ± 4a | 3.70 ± 0.19a | ND |
| WC | 0 | ND | 0 | 0.33 ± 0.13c |
All data are averages (±SEM) from five, or from ten (dry weight) independent plants. *Small nodules are of length <2 mm and include both pink and white nodules. a,b,cStudent's t test was performed within each experiment and letters indicate significant difference (P ≤ 0.01). WC, water control is uninoculated; ND not determined.
To determine survival of the GSH-deficient mutant in the pea rhizosphere, a low number of bacteria (103 cfu) were introduced onto seedling roots and the total bacteria determined after 7 days’ plant growth. When individual LMB599 and wild-type Rlv3841 strains were used to inoculate a sterile pea rhizosphere, the population density of the mutant strain recovered is only 35% that of wild type (Fig. 3). Competition between the GSH-deficient mutant and wild type for growth in the pea rhizosphere was measured by inoculating pea roots with both bacterial strains together. When inoculated in equal ratios, LMB599 accounts for only 3% of bacteria recovered (t test; P ≤ 0.001). Even when strain LMB599 was inoculated at a 10-fold excess over wild type, it accounts for only 36% of the total bacteria recovered (Fig. 3).
Figure 3.
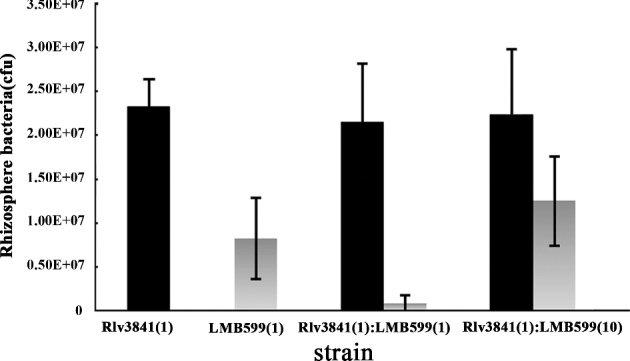
Colonisation of the rhizosphere of pea plants by wild-type Rlv3841 (black) and GSH-deficient mutant LMB599 (grey), inoculated individually and together. Inoculation ratios are given on the x-axis, with 1 corresponding to 103 cfu. Rhizosphere bacteria is the average number of bacteria (cfu) recovered per plant (±SEM). Number of replicates = 10.
DISCUSSION
In Rhizobium etli, the presence of gshB, encoding GSH synthetase, is essential for growth on glutamine as the sole source of carbon and nitrogen (Taté et al.2012). In GSH-deficient strains of this organism, glutamine transport was reported to be the defective step in glutamine utilisation (Taté et al.2012). Here, we focus on a GSH-deficient strain of R. leguminosarum that has an impaired growth phenotype and examine the uptake of various carbon compounds. The R. leguminosarum gshB mutant grows poorly on a variety of carbon sources including a sugar (glucose), an organic acid (succinate) and amino acids (glutamine, glutamate, histidine, alanine and asparagine). The growth defect is rescued either with a plasmid encoding a functional gshB or, on glucose and succinate and, to some extent, histidine and alanine, by addition of exogenous GSH. GSH, either produced inside the bacterial cell or added externally, affects uptake and/or utilisation of these compounds in R. leguminosarum.
In the GSH-deficient mutant, transport assays were used to determine the rate of uptake of three compounds; AIB (an amino acid analogue), glucose and succinate. Since the rate of uptake of non-metabolisable AIB is reduced to a similar extent to those of glucose and succinate (60%–69%), GSH may be required for the full activity of many different transport systems. It is noteworthy that rhizobia have a high number of ABC transport systems including (but not exhaustively) three systems for uptake of glucose (Untiet et al.2013) and Aap/Bra for amino acids (including AIB) (Hosie et al.2002).
Since the uptake of many of the carbon and nitrogen sources examined is dependent on ABC transport systems, we thought it possible that the PTSNTR system, which is a global regulator of many transporters particularly ABC transport systems (Prell et al.2012; Untiet et al.2013), may exert its effect via GSH. Strain PtsP107 shows significantly reduced transport of AIB; however, exogenously added GSH is unable to rescue this deficiency in transport. This indicates that GSH does not regulate amino acid uptake in the PTSNTR system, and there must be another mechanism(s) affecting transport.
Rhizobium leguminosarum has two general amino acid permeases, Aap and Bra, which are both broad specificity amino acid transport systems (Hosie et al.2002) and able to transport AIB (assayed here) and, more importantly, in the context of testing it is important in the R. leguminosarum system, glutamine. We determined whether transcription of these two-transport systems is regulated by GSH. In the R. leguminosarum gshB mutant, expression of all Bra system genes tested (braC, braD, braE and braG) is >2-fold downregulated compared to wild-type; however, the expression of Aap system genes aapJ and aapQ is essentially unchanged. This shows that GSH positively regulates the transcription of bra but not aap genes. Interestingly, the converse was observed in R. etli, where the essential role of GSH in glutamine uptake was through regulation of the Aap transport system and the Bra system was found to be dispensable (Taté et al.2012). Thus, lack of GSH reduces transcription of some, but not all, R. leguminosarum transport operons suggesting that GSH does not globally alter transcription of transport genes by means of a single mechanism. The absence of a general transcriptional effect suggests that GSH may directly or indirectly, for example, via redox changes, alter the assembly or activity of a broad range of transport systems by a combination, or even an unknown, mechanism(s). Lack of a single mechanism may be the reason for the different patterns of growth seen on amino acids in both wild-type and mutant strains when exogenous GSH is added (Fig. 1). The fact that only on glutamine is the growth of wild-type Rlv3841 poorer in the presence of additional GSH than in its absence, suggests that glutamine transport and/or metabolism is adversely affected by excess GSH. Additional GSH restores growth to the mutant only on glutamate, histidine and alanine (not glutamine), while the plasmid-complemented mutant LMB675 grows even better than wild-type on asparagine. Perhaps in this last case there is increased transport of asparagine, possibly due to increased level of GSH (on a multicopy plasmid expression of gstB is likely to be higher than from a single chromosomal copy), leading to increased expression of the Bra transporter.
In symbiosis, a R. leguminosarum GSH-deficient mutant shows an approx. 50% reduction in the dry weight of nodulated plants and diminished nitrogen-fixing capacity (reduced to approx. 50%), although the total number of nodules was unchanged. During nodule formation and symbiosis, legume roots produce antioxidants including GSH (Becana et al.2010; Matamoros et al.2015). As growth defects in the free-living gshB mutant are rescued by exogenous GSH, it might have been expected that GSH from the plant would be able to perform the same role in symbiotic interactions. However, any antioxidants, including GSH, produced by pea roots are clearly unable to rescue the gshB mutation in symbiosis under these conditions as a significant feature of our study is the large the proportion (approx. 75%) of undeveloped and ineffective nodules formed by the mutant strain. This may be due to early senescence, which was observed in the nodules from gshB mutants of R. tropici, Ensifer meliloti and R. etli (Harrison et al.2005; Muglia et al.2008). In R. tropici and E. meliloti gshB mutants, the total number of nodules formed was found to be similar to that from equivalent wild-type strains (Riccillo et al.2000; Harrison et al.2005), which is also the case for R. leguminosarum in this study. Moreover, the reduced ability of the R. leguminosarum gshB mutant to grow effectively in a sterile rhizosphere and to compete with its wild-type parent in colonising plant roots shows that bacterial GSH production is important for adaption to the microenvironment of host plants.
An inability to effectively colonise pea roots, poor nodulation, reduced dry weight of inoculated plants and diminished nitrogen fixation shows that normal GSH metabolism is essential for the whole symbiotic process in R. leguminosarum. These observations, together with the poor competitive ability to nodulate plants shown by GSH-deficient mutants of E. meliloti (Harrison et al.2005) and R. etli (Taté et al.2012), suggest that lack of bacterial GSH production has a profound influence on symbiosis in many different rhizobia.
FUNDING
This work was supported by the Biotechnology and Biological Sciences Research Council [grant numbers BB/J007749/1, BB/J007749/2] and the Fundamental Research Funds for the Central Universities, South-Central University for Nationalities [grant numbers CZW15023, CZW15112] to GC.
The research materials supporting this publication can be accessed by contacting philip.poole@plants.ox.ac.uk.
Conflict of interest. None declared.
REFERENCES
- Allaway D, Lodwig E, Crompton LA et al. . Identification of alanine dehydrogenase and its role in mixed secretion of ammonium and alanine by pea bacteroids. Mol Microbiol 2000;36:508–15. [DOI] [PubMed] [Google Scholar]
- Barr M, East AK, Leonard M et al. . In vivo expression technology (IVET) selection of genes of Rhizobium leguminosarum biovar viciae A34 expressed in the rhizosphere. FEMS Microbiol Lett 2008;282:219–27. [DOI] [PubMed] [Google Scholar]
- Becana M, Matamoros MA, Udvardi M et al. . Recent insights into antioxidant defenses of legume root nodules. New Phytol 2010;188:960–76. [DOI] [PubMed] [Google Scholar]
- Beringer JE. R factor transfer in Rhizobium leguminosarum. J Gen Microbiol 1974;84:188–98. [DOI] [PubMed] [Google Scholar]
- Clarke VC, Loughlin PC, Gavrin A et al. . Proteomic analysis of the soybean symbiosome identifies new symbiotic proteins. Mol Cell Proteomics 2015;1414:1301–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figurski DH, Helinski DR. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. P Natl Acad Sci USA 1979;76:1648–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison J, Jamet A, Muglia CI et al. . Glutathione plays a fundamental role in growth and symbiotic capacity of Sinorhizobium meliloti. J Bacteriol 2005;187:168–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosie AH, Allaway D, Galloway CS. et al. Rhizobium leguminosarum has a second general amino acid permease with unusually broad substrate specificity and high similarity to branched-chain amino acid transporters (Bra/LIV) of the ABC family. J Bacteriol 2002;184:4071–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston AW, Beringer JE. Identification of the rhizobium strains in pea root nodules using genetic markers. J Gen Microbiol 1975;87:343–50. [DOI] [PubMed] [Google Scholar]
- Karunakaran R, Ebert K, Harvey S et al. . Thiamine is synthesized by a salvage pathway in Rhizobium leguminosarum bv. viciae strain 3841. J Bacteriol 2006;188:6661–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karunakaran R, Haag AF, East AK et al. . BacA is essential for bacteroid development in nodules of galegoid, but not phaseoloid, legumes. J Bacteriol 2010;192:2920–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long SR. Rhizobium symbioses: nod factors in perspective. Plant Cell 1996;8:1885–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matamoros M, Saiz A, Penuelas M et al. . Function of glutathione peroxidases in legume root nodules. 2015J Exp Bot;66:2979–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muglia C, Comai G, Spegazzini E et al. . Glutathione produced by Rhizobium tropici is important to prevent early senescence in common bean nodules. FEMS Microbiol Lett 2008;286:191–98. [DOI] [PubMed] [Google Scholar]
- Mulley G, White JP, Karunakaran R et al. . Mutation of GOGAT prevents pea bacteroid formation and N2 fixation by globally down-regulating transport of organic nitrogen sources. Mol Microbiol 2011;80:149–67. [DOI] [PubMed] [Google Scholar]
- Oldroyd GED, Murray JD, Poole PS et al. . The rules of engagement in the legume-rhizobial symbiosis. Ann Rev Genet 2011;45:119–44. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST(c)) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 2002;30:E36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole PS, Blyth A, Reid C et al. . myo-Inositol catabolism and catabolite regulation in Rhizobium leguminosarum bv. viciae. Microbiology 1994;140:2787–95. [Google Scholar]
- Prell J, Bourdès A, Karunakaran R et al. . Pathway of γ-aminobutyrate (GABA) metabolism in Rhizobium leguminosarum 3841 and its role in symbiosis. J Bacteriol 2009;191:2177–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prell J, Mulley G, Haufe F et al. . The PTS(Ntr) system globally regulates ATP-dependent transporters in Rhizobium leguminosarum. Mol Microbiol 2012;84:117–29. [DOI] [PubMed] [Google Scholar]
- Reid CJ, Poole PS. Roles of DctA and DctB in signal detection by the dicarboxylic acid transport system of Rhizobium leguminosarum. J Bacteriol 1998;180:2660–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro CW, Alloing G, Mandon K et al. . Redox regulation of differentiation in symbiotic nitrogen fixation. Biochim Biophys Acta 2015;1850:1469–78. [DOI] [PubMed] [Google Scholar]
- Riccillo PM, Muglia CI, de Bruijn FJ et al. . Glutathione is involved in environmental stress responses in Rhizobium tropici, including acid tolerance. J Bacteriol 2000;182:1748–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taté R, Cermola M, Riccio A et al. . Glutathione is required by Rhizobium etli for glutamine utilization and symbiotic effectiveness. Mol Plant Microbe In 2012;25:331–40. [DOI] [PubMed] [Google Scholar]
- Udvardi M, Poole PS. Transport and metabolism in legume-rhizobia symbioses. Annu Rev Plant Biol 2013;64:781–805. [DOI] [PubMed] [Google Scholar]
- Untiet V, Karanukaran R, Krämer M et al. . ABC transport is inactivated by the PTSNtr under potassium limitation in Rhizobium leguminosarum 3841. PLoS One 2013;8:e64682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walshaw DL, Poole PS. The general L-amino acid permease of Rhizobium leguminosarum is an ABC uptake system that also influences efflux of solutes. Mol Microbiol 1996;21:1239–52. [DOI] [PubMed] [Google Scholar]