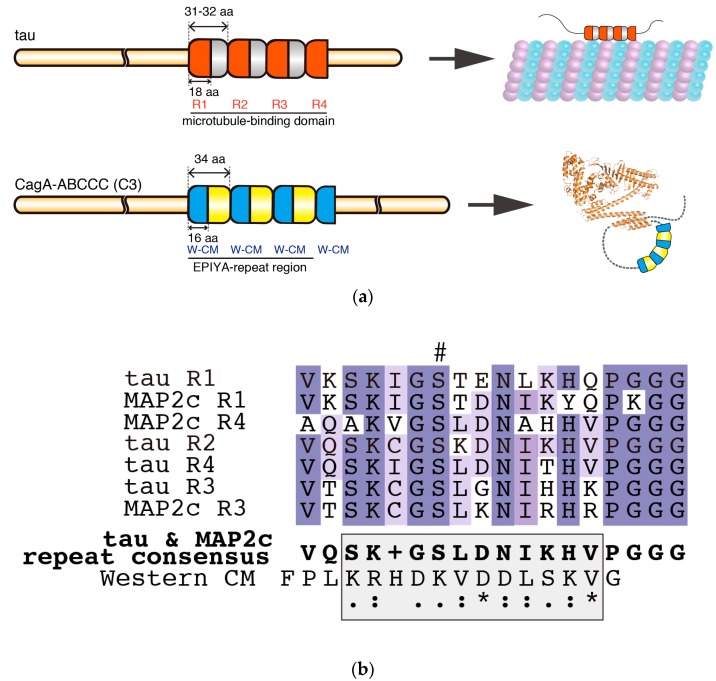
Figure 3.
Comparison of the intrinsically disordered C-terminal repeat regions of tau and CagA. (a) The repeat regions of tau and CagA have similar segmental organizations. The 2N4R tau (RefSeq Accession: NP_005901.2) has a microtubule-binding domain that is comprised of four degenerate repeat segments. Each of these 31–32 amino-acid repeat segments contains a semi-conserved 18 amino-acid core repeat termed R1–R4. The microtubule-binding domain binds and stabilizes microtubules; however, tau itself remains intrinsically disordered. R1–R4 of tau are targets of PAR1b and their phosphorylation results in dissociation of tau from microtubules. In comparison, the EPIYA-repeat region of ABCCC-type CagA (NCTC11637 strain) is comprised of three highly conserved 34 amino-acid EPIYA-C segments, each of which contains a fully conserved 16 amino-acid Western CM motif. Another fully conserved Western CM motif also exists immediately distal to the third repeat of the EPIYA-C segments. The CM motifs are located in the C-terminal tail of CagA, which forms a flexible lariat loop; (b) Multiple sequence alignment of tau and MAP2c (RefSeq Accession: NP_114033-2) R1-R4 repeats revealed a consensus sequence that showed weak similarity (shaded box) with the Western CM sequence. Hash denotes phosphorylation target site of PAR1b. Multiple sequence alignment and consensus sequence were generated by Clustal Omega [78].