Abstract
Sal-like protein 4 (SALL4), an embryonic stem cell transcriptional regulator, is re-expressed by an unknown mechanism in poor prognosis hepatocellular carcinoma (HCC), often associated with chronic hepatitis B virus (HBV) infection. Herein, we investigated the mechanism of SALL4 re-expression in HBV-related HCCs. We performed bisulfite sequencing PCR of genomic DNA isolated from HBV-related HCCs and HBV replicating cells, and examined DNA methylation of a CpG island located downstream from SALL4 transcriptional start site (TSS). HBV-related HCCs expressing increased SALL4 exhibited demethylation of specific CpG sites downstream of SALL4 TSS. Similarly, SALL4 re-expression and demethylation of these CpGs was observed in HBV replicating cells. SALL4 is also re-expressed in poor prognosis HCCs of other etiologies. Indeed, increased SALL4 expression in hepatitis C virus-related HCCs correlated with demethylation of these CpG sites. To understand how CpG demethylation downstream of SALL4 TSS regulates SALL4 transcription, we quantified by chromatin immunoprecipitation (ChIP) assays RNA polymerase II occupancy of SALL4 gene, as a function of HBV replication. In absence of HBV replication, RNA polymerase II associated with SALL4 exon1. By contrast, in HBV replicating cells RNA polymerase II occupancy of all SALL4 exons increased, suggesting CpG demethylation downstream from SALL4 TSS influences SALL4 transcriptional elongation. Intriguingly, demethylated CpGs downstream from SALL4 TSS are within binding sites of octamer-binding transcription factor 4 (OCT4) and signal transducer and activator of transcription3 (STAT3). ChIP assays confirmed occupancy of these sites by OCT4 and STAT3 in HBV replicating cells, and sequential ChIP assays demonstrated co-occupancy with chromatin remodeling BRG1/Brahma-associated factors. BRG1 knockdown reduced SALL4 expression, whereas BRG1 overexpression increased SALL4 transcription in HBV replicating cells. We conclude demethylation of CpGs located within OCT4 and STAT3 cis-acting elements, downstream of SALL4 TSS, enables OCT4 and STAT3 binding, recruitment of BRG1, and enhanced RNA polymerase II elongation and SALL4 transcription.
Keywords: SALL4, DNA methylation, HBV Hepatocellular Carcinoma (HCC), RNA Polymerase II pausing, BAF complex
INTRODUCTION
Liver cancer has the most rapidly growing mortality rate in the U.S., relative to other cancers, and it is the 3rd cause of death by cancer worldwide32. Investigations of genes involved in liver cancer have focused on Sal-like protein 4 (SALL4), a marker of embryonic stem cells41 and a prognostic marker in liver cancer14, 27, 37. SALL4 is an oncofetal protein expressed at high levels in fetal liver while its expression is silenced in adult hepatocytes26. Recent studies have shown increased expression of SALL4 in poor survival HCCs14. SALL4 is re-expressed in 95 of 171 HBV-related HCC patients (55.6%) with unfavorable prognosis37. SALL4 expression exhibits a strong positive correlation with expression of EpCAM, hepatic stem cell markers, and AFP29, 36, 38. Importantly, SALL4 expression enhanced formation of spheroids and invasion capacities of HCC-derived cell lines, while SALL4 knockdown led to reduction in expression of these markers, and attenuation of invasion capacity38. These findings support a role for SALL4 in hepatocarcinogenesis associated with unfavorable prognosis. Accordingly, SALL4 has been proposed as a novel therapeutic target for liver cancer, although transcription factors are not effective druggable targets23, 27.
Given the important role of SALL4 in liver cancer, we aimed to investigate the molecular mechanism that enables re-expression of the SALL4 gene in liver tumors. The SALL4 promoter contains high affinity signal transducer and activator of transcription 3 (STAT3) binding sites, and down-regulation of STAT3 significantly reduced SALL4 expression3. In embryonic stem cells (ESCs) SALL4 expression was dramatically enhanced by overexpression of octamer-binding transcription factor 4 (OCT4)34. Furthermore, in ESCs the BRG1/Brahma-associated factors (BAFs) complex occupies the SALL4 promoter, suggesting the BAF complex may also have a regulatory role in SALL4 re-expression in tumors11. Significantly, the activity and expression of SALL4 is epigenetically regulated. For example, the histone deacetylase inhibitor successfully suppressed proliferation of SALL4-positive hepatocellular carcinoma cells, while the DNA methylation inhibitor 5-azacytidine (5AZA) specifically reversed repressive effect of SALL4 on its own expression as well as on SALL4 target genes35. SALL4 level was reduced by overexpression of miR-107, and the inverse was observed when miR-107 was knocked-down9. In addition, H3K27 demethylase Utx directly regulates re-expression of SALL4 in somatic and germ cell reprogramming22 while DNA (cytosine-5)-methyltransferase 3-like (DNMT3L) is required to suppress SALL4 splicing variant SALL4B19. Genome-wide DNA methylation analysis in ESCs revealed SALL4 was hypo-methylated in stem cell-specific differentially methylated regions (SS-DMRs)25.
However, little is known about the molecular mechanism regulating re-expression of SALL4 in liver tumors. Since SALL4 is re-expressed in nearly 50% of HBV-related HCCs37, in this study we investigated how HBV infection regulates SALL4 expression, employing in vitro HBV replication17 and infection models24. Herein, we identify a cluster of CpG sites that become demethylated in HBV replicating cells and HBV-related liver tumors. These CpG sites, located downstream from the TSS of SALL4, upon DNA demethylation enable STAT3 and OCT4 binding, recruitment of chromatin remodeling complex containing BRG1, and increased RNA Polymerase II association with SALL4 chromatin, resulting in enhanced SALL4 transcription. Furthermore, we provide evidence that this mechanism of SALL4 re-expression is also functional in HCC of other etiologies, including HCV-related HCCs, and in human liver cancer cell lines expressing elevated SALL4 mRNA37.
RESULTS
DNA demethylation of specific CpG sites located downstream of SALL4 TSS in HBV-related HCCs
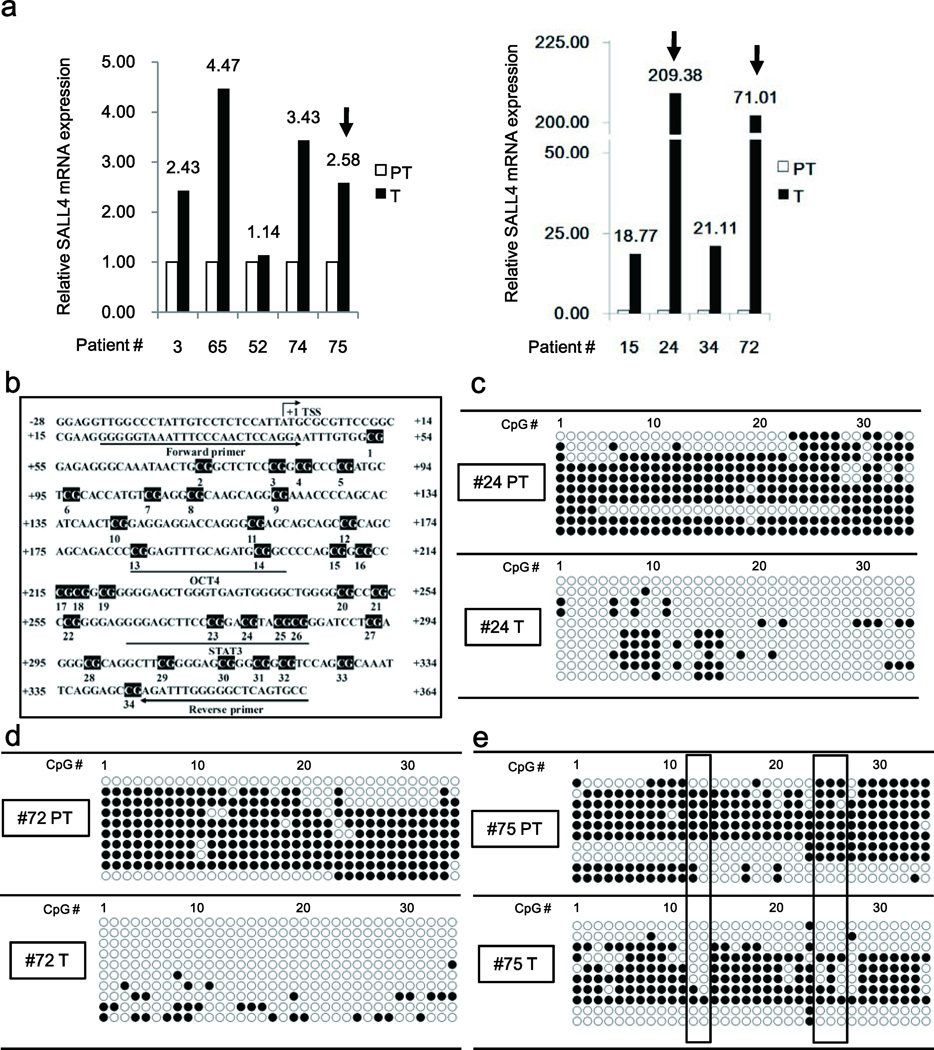
Our studies demonstrated hepatitis B virus X protein induces EpCAM expression via active DNA demethylation7. To examine whether SALL4 transcription is also regulated by HBV via a similar DNA demethylation mechanism, we performed sodium bisulfate sequencing PCR of DNA isolated from HBV-related HCCs (T) and paired peritumor liver tissue (PT). Firstly, we quantified SALL4 expression employing seventeen clinical samples. Of them, five patients showed slightly elevated mRNA level (left panel, Fig. 1a) and four patients showed significantly increased SALL4 mRNA levels (right panel, Fig. 1a), compared to PT tissue. SALL4 mRNA expression was either not upregulated or reduced in the remaining eight patients (Supplementary Fig. 1a).
Figure 1. SALL4 expression is associated with DNA demethylation in a subgroup of HBV-related HCCs.
(a) PCR quantification of SALL4 mRNA in HBV-related HCCs with mild (Left panel) and extreme increase (Right panel) of SALL4 expression. PT, peritumor; T, tumor. (b) Mapping of CpG dinucleotides in human SALL4 gene. The transcriptional start site (TSS), numbered CpG sites, OCT4 and STAT3 binding sites, and the position of bisulfite sequencing primers are indicated. (c–e) Bisulfite sequencing PCR of SALL4 clones, using DNA from patients #24 (c), #72 (d), and #75 (e), as indicated in Fig. 1a. Open and closed circles denote unmethylated and methylated states, respectively.
Transcription factor binding sites (TFBS) are usually located 1–2 kb upstream of the transcription start site (TSS)4. Accordingly, we used the 5’ flanking sequence of SALL4 spanning 2Kb upstream and 500 bp downstream from TSS for CpG island and TFBS prediction (Fig. 1b). Sequence analysis revealed 34 CpG sites and multiple TFBS, including STAT3 and OCT4 binding sites, located downstream from the SALL4 TSS (Fig. 1b).
Next, we used samples from patients #24 and #72, exhibiting high expression of SALL4 mRNA, 209-fold and 71-fold respectively, and patient #75 with small 2.6-fold induction of SALL4 mRNA (Fig. 1a). Sodium bisulfite sequencing PCR demonstrated vast DNA demethylation in T sample #24 (Fig. 1c) and #72 (Fig. 1d). Interestingly, these T samples exhibited high expression of SALL4 mRNA (Fig. 1a). By contrast, comparison of DNA methylation pattern between T vs. PT tissue from patient #75, exhibiting moderate SALL4 mRNA expression (Fig. 1a), showed that CpG sites 12–13 and 24–27 were demethylated in the tumor, suggesting that demethylation at those sites is functionally important for SALL4 re-expression (Fig. 1e).
HBV infection induces DNA demethylation of specific SALL4 CpG sites
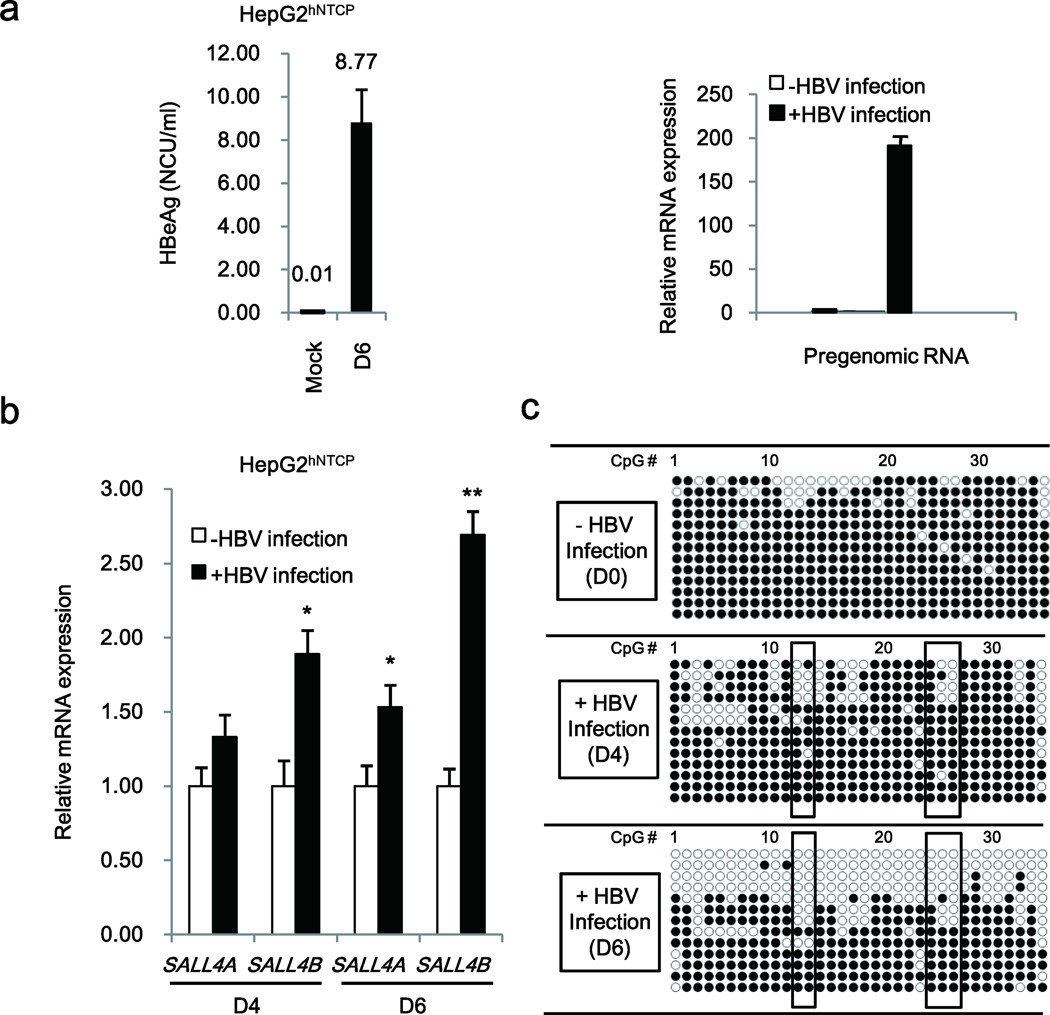
Since the observed DNA demethylation of CpG sites located downstream from SALL4 TSS in HBV-related HCCs occurs in the context of chronic HBV infection, we investigated whether this SALL4 CpG site demethylation also occurs upon HBV infection of hepatocytes in vitro. We employed the recently developed HBV infection model of the HepG2hNTCP cell line24. HepG2hNTCP cells express sodium taurocholate co-transporting peptide (NTCP) that specifically binds HBV preS1 protein and can be directly infected by purified HBV virus. Quantification of viral HBeAg and pregenomic RNA were used to detect HBV infection in HepG2hNTCP cells (Fig. 2a). Indeed, HBV-infected HepG2hNTCP cells showed increased expression of SALL4 splicing variants SALL4A and SALL4B (Fig. 2b). Interestingly, sodium bisulfite sequencing analyses of DNA isolated from cells infected with HBV for 4 days and 6 days displayed a progressive increase in DNA demethylation of specific CpG sites located downstream of SALL4 TSS (Fig. 2c). Statistical analysis by QUMA16 displayed significant hypomethylation at CpG sites 12–14 and 24–26 (Supplementary Fig. 2a). These CpG sites were also found hypomethylated in HBV-related HCCs (Fig. 1) and interestingly, they are located within or adjacent to putative binding sites for OCT4 and STAT3 (Fig. 2c).
Figure 2. HBV infection induces DNA demethylation of SALL4 in HepG2hNTCP cell line.
(a) Quantification of secreted HBeAg by ELISA (Left panel), and quantification of pregenomic RNA by QPCR (Right panel) after HBV infection for 6 days. (b) Quantification of SALL4 gene expression (SALL4A and SALL4B slicing variants) by QPCR after HBV infection for 4 and 6 days (D4, D6). (c) Bisulfite sequencing PCR results of SALL4 clones, using DNA from HepG2hNTCP cells without (−) HBV infection (D0) or with (+) HBV infection at 4 and 6 days (D4 and D6). Open and closed circles denote unmethylated and methylated states, respectively. Error bars denote S.D. *P< 0.05; **P< 0.01.
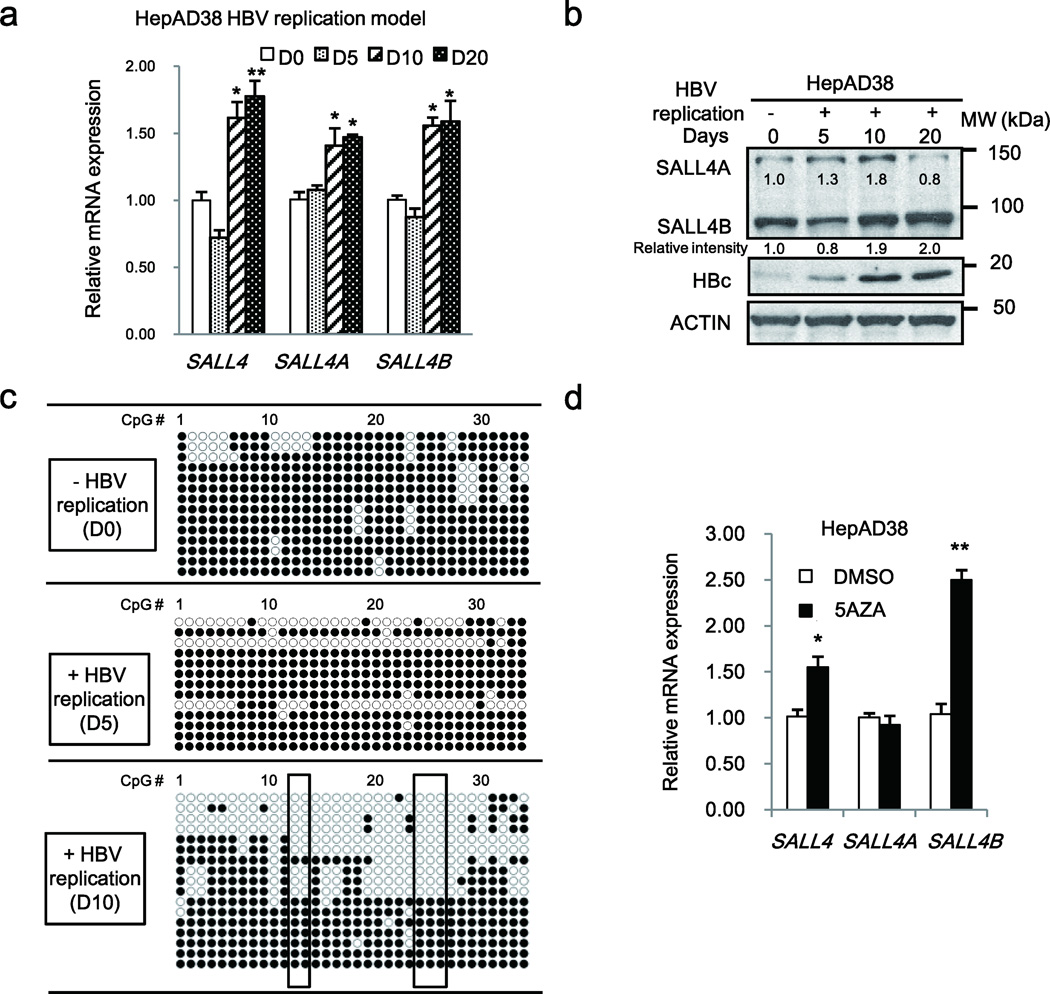
To further verify these DNA demethylation results derived from clinical samples and the HepG2hNTCP HBV infection model, we also employed the HepAD38 cell line, a cellular model for HBV replication17. HepAD38 cells, derived from the HepG2 human liver cancer cell line, contain an integrated copy of the HBV genome under control of the Tet-off promoter, and HBV replication is initiated by tetracycline removal17. Under conditions of HBV replication by tetracycline removal for 5 days (D5), 10 days (D10) and 20 days (D20), monitored by increased protein levels of viral HBc antigen (Fig. 3b), qRT-PCR results showed significant mRNA induction of SALL4A and SALL4B splicing variants at D10 and D20 (Fig. 3a). Protein expression results by immunoblotting confirmed this mRNA increase (Fig. 3b). Interestingly, significant DNA demethylation was observed at D10 and D20 of HBV replication but not on D5, compared to cells without HBV replication (Fig. 3c and Supplementary Fig. 3a and b). Importantly, in D10 and D20 of HBV replicating HepAD38 cells, CpG sites 12–14 and 24–27 were hypomethylated, agreeing with results derived from HBV infected HepG2hNTCP (Fig. 2) and HBV-related HCCs (Fig. 1). Thus, we interpret these results to indicate that DNA demethylation of CpG sites 12–14 and 24–27 downstream of the TSS is functionally important in SALL4 re-expression.
Figure 3. HBV replication induces DNA demethylation of SALL4 in HepAD38 cell line.
(a) PCR quantification of SALL4 mRNA in HepAD38 cells grown without (−) HBV replication (D0) or with (+) HBV replication by tetracycline removal for 5, 10, 20 days (D5–D20). Results are from three independent RNA isolations performed in identical triplicates. Error bars denote S.D. *P< 0.05; **P< 0.01. (b) Immunoblot of SALL4 protein in HepAD38 cells −/+ HBV replication by tetracycline removal for D0–D20. (c) Bisulfite sequencing PCR results of SALL4 clones, using DNA from HepAD38 cells −/+ HBV replication by tetracycline removal for D0–D10. Open and closed circles denote unmethylated and methylated states, respectively. (d) QPCR of SALL4 mRNA following treatment with 10 µM 5-Aza-2-deoxycytidine (5AZA) or DMSO (vehicle control) for 48hr. Results are from three independent RNA isolations performed in identical triplicates. Error bars denote S.D. *P< 0.05; **P< 0.01.
To link SALL4 mRNA expression to DNA demethylation, we treated HepAD38 cells with 5AZA, an inhibitor of DNA methyltransferases (DNMTs). Indeed, SALL4 mRNA levels were elevated by treatment with 5AZA in the absence of HBV replication (Fig. 3d). These results indicate that DNA demethylation is required for SALL4 re-expression in hepatocytes, involving a passive mechanism.
SALL4 re-expression in liver cancer cell lines and HCV-related HCCs via DNA demethylation
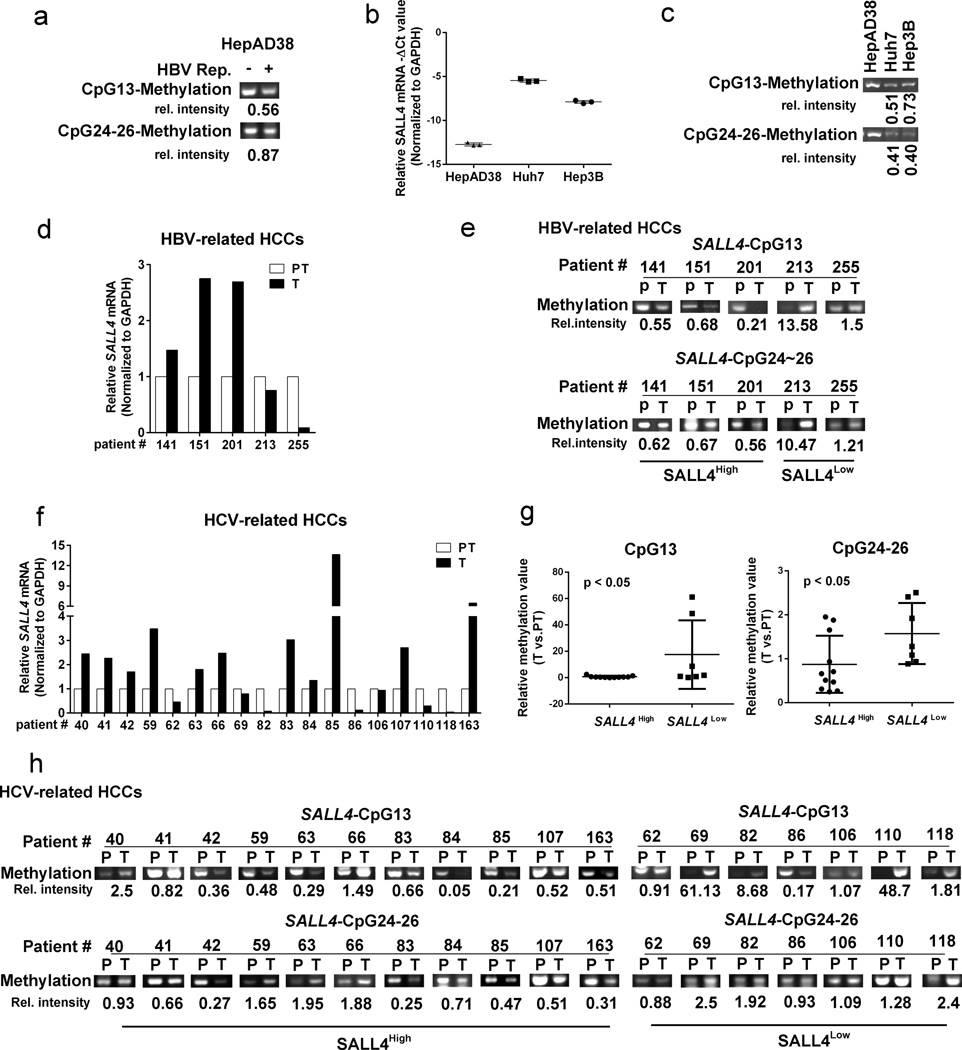
To further confirm this mechanism of SALL4 re-expression observed in differentiated hepatocytes, we developed a methylation specific PCR (MSP) assay10. The MSP assay involves modification of DNA by sodium bisulfite to convert unmethylated cytosines to uracil, followed by PCR amplification with primers specific for the methylated DNA10. Specifically, employing the HepAD38 cell line as our positive control, which exhibits HBV replication-dependent DNA demethylation (Fig. 3), we designed primers to detect the methylation state of CpG sites associated with OCT4 and STAT3 binding sites at positions CpG13 and CpG24–26, downstream from SALL4 TSS. In agreement with the HepAD38 bisulfite sequencing data (Fig. 3c) our MSP primers exhibited reduced methylation of CpG13 and CpG24–26 sites, employing genomic DNA isolated from HepAD38 cells on D10 of HBV replication (Fig. 4a).
Figure 4. SALL4 re-expression in liver cancer cell lines and HCV-related HCCs via DNA demethylation.
(a) Methylation-specific PCR (MSP) assay for SALL4 CpG site 13 and CpG sites 24–26, employing genomic DNA from HepAD38 cells grown without (−) and with (+) HBV replication for 10 days, analyzed 2% agarose gel electrophoresis. Relative intensity quantified by ImageJ software is ratio of +/− HBV replication. (b) QRT-PCR of SALL4 mRNA expression employing RNA from HepAD38 cells grown in the absence of HBV replication, Huh7 and Hep3B cell lines. Data normalized to GAPDH. −ΔCt values are shown. (c) MSP assay for CpG site 13 and CpG24–26 using genomic DNA from HepAD38 (without HBV replication), Huh7 and Hep3B cells, analyzed by 2% agarose gel electrophoresis. Relative intensity quantified by ImageJ software is ratio of signal from Huh7 or Hep3B to HepAD38 cells. (d) and (f) QRT-PCR of SALL4 mRNA expression in HBV- and HCV- related liver tumors (T) vs. peritumoral tissue (PT); (e) and (h) MSP assay for methylation of CpG13 and CpG24–26 sites using genomic DNA of HBV- and HCV- related HCCs; relative intensity is ratio of T/PT. (g) Statistical analysis for methylation status of SALL4 CpG13 and CpG24–26 sites in patient samples with SALL4High and SALL4low mRNA expression, using unpaired t test; P < 0.05.
To further validate the MSP assay, we analyzed human liver cancer cell lines shown in earlier studies37 to re-express SALL4. Indeed, in comparison to HepAD38 cells not replicating the virus, SALL4 is highly re-expressed in Huh7 and Hep3B cell lines (Fig. 4b), in agreement with earlier studies by others37. Employing genomic DNA from these cell lines, including genomic DNA from HepAD38 cells without HBV replication, we demonstrate that indeed the designed PCR primers distinguish methylated vs. unmethylated DNA at those CpG sites (Fig. 4c). More importantly, the results demonstrate that HCC cell lines that re-express SALL4 also exhibit demethylation of these CpG sites downstream of the SALL4 TSS, suggesting that this mechanism is operational in liver cancer independent of HBV infection (Fig. 4c).
Next, we examined SALL4 expression in T vs. PT tissue from a small cohort of chronic HBV infected patients with HCC, and another cohort of HCV-associated HCCs. Three out of five HBV-related HCCs and 11 out of 18 HCV-related HCCs were positive for SALL4 expression (Fig. 4d and Fig. 4f). Indeed, our MSP analyses demonstrate that the state of DNA methylation at the SALL4 regulatory region differed between T and PT tissue in SALL4 expressing tumors (Fig. 4e and h). HCV-related HCCs expressing SALL4 exhibited a statistically significant reduction (p<0.05) in DNA methylation at CpG positions 13 and 24–26 (Fig. 4g). Importantly, HCCs expressing SALL4 exhibit demethylation of at least one of the CpG sites associated with OCT4 or STAT3 binding sites (Fig. 4e and h). We conclude, DNA demethylation of CpG sites associated with OCT4 and/or STAT3 binding sites located downstream from the SALL4 TSS, is functionally relevant for re-expression of SALL4 in hepatocarcinogenesis.
CpG site methylation downstream of SALL4 TSS interferes with RNA polymerase II elongation
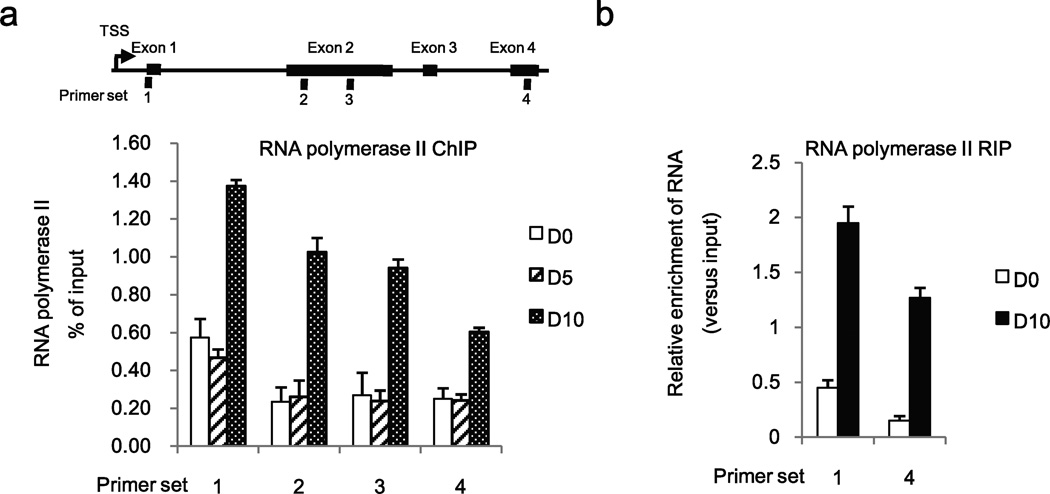
Since DNA methylation at coding region boundaries6 negatively influences RNA polymerase II binding to chromatin20 and elongation rate33, we performed ChIP assays to quantify RNA polymerase II occupancy in SALL4 regulatory region located in boundary region of exon 1, using primer set 1 (Fig. 5a). In addition, in order to monitor association of RNA polymerase II with chromatin of exons 2–4, as a function of HBV replication, we used primer sets 2–4 (Fig. 5a). Increased association (50%) of RNA polymerase II was detected by ChIP assays with exon 1 on D0, i.e., without HBV replication, in comparison to exons 2–4 (Fig. 5a). Similarly, at D5 of HBV replication, when CpG sites in boundary of exon 1 were still methylated (Fig. 3c), RNA polymerase II associated primarily with exon 1(Fig. 5a). By contrast, at D10 of HBV replication, ChIP assays demonstrated increased association of RNA polymerase II with the chromatin of all SALL4 gene exons (Fig. 5a). RNA immunoprecipitation (RIP) assays employing RNA polymerase II antibody confirmed that RNA polymerase II associated with significantly increased SALL4 transcript at D10 of HBV replication, corresponding to both exon1 and exon 4 (Fig. 5b). By contrast, in the absence of HBV replication (D0), RIP assays show that RNA Polymerase II was primarily in association with RNA corresponding to exon 1. Taken together, we interpret these result to mean that in the absence of HBV replication (D0), or when CpG sites located in the boundary of exon 1 were methylated, as on D5 of HBV replication (Fig. 5a), RNA polymerase II was paused at the region of the methylated CpG sites. Upon demethylation of these CpG sites RNA polymerase II resumed transcription elongation.
Figure 5. DNA methylation downstream of SALL4 TSS interferes with RNA polymerase II elongation.
(a) Diagram of SALL4 gene. TSS and exon1–4 are shown. ChIP assays with RNA polymerase II antibody, employing HepAD38 cells grown −/+ HBV replication by tetracycline removal for D0–D10, and SALL4 primers spanning different exons, as indicated. (b) RIP assays with RNA polymerase II antibody, employing HepAD38 cells grown −/+ HBV replication by tetracycline removal for D0 and D10, and SALL4 primers spanning exons 1 and 4, as indicated in (a).
HBV replication-induced DNA demethylation facilitates binding and interaction of STAT3, OCT4 and BRG1
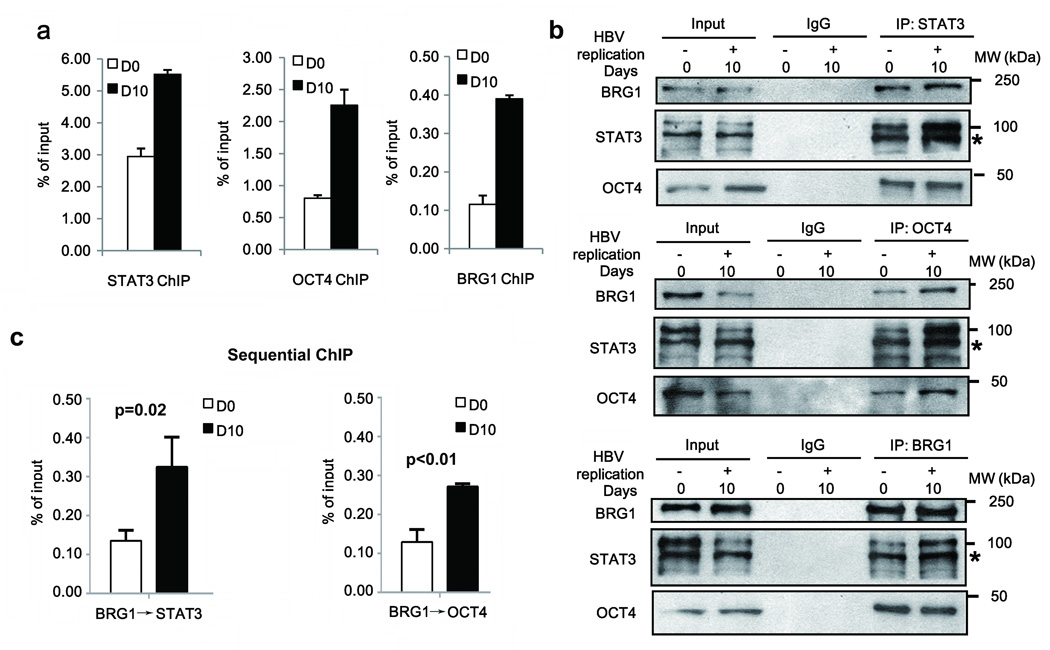
Interestingly, the CpG sites that were demethylated both in HBV-related HCCs and in HBV infected and replicating cells are located within or adjacent to putative transcription factor binding sites. CpG sites 13 and 14 are located in an OCT4 binding site, and CpG sites 23–26 in a STAT3 binding site. Earlier studies demonstrated that STAT33 and OCT434 bind to their cis-acting sequences located in SALL4 promoter region and activate SALL4 expression in ESCs. Interestingly, our results suggest that DNA methylation of STAT3 and OCT4 sites located downstream of the SALL4 TSS, prevent SALL4 transcription in differentiated hepatocytes. To directly demonstrate whether DNA demethylation affects binding of OCT4 and STAT3 at their respective binding sites downstream from the TSS, we carried out ChIP assays, using OCT4 and STAT3 antibodies. ChIP results show increased binding of STAT3 and OCT4 (Fig. 6a) at those sites in the presence of HBV replication at D10, i.e. when the CpG sites become demethylated (Fig. 3c).
Figure 6. HBV replication enables binding and interaction of STAT3, OCT4 and BRG1 in SALL4 regulatory region.
(a) ChIP assays with STAT3, OCT4, and BRG1 antibodies, as indicated, employing HepAD38 cells grown −/+ HBV replication by tetracycline removal for D0 and D10, and SALL4 primers spanning CpG sites shown in Fig.1b. Data were quantified as % of input. (b) Co-immunoprecipitations (co-IPs) of STAT3, OCT4 and BRG1 using indicated antibodies, employing extracts from HepAD38 cells grown −/+ HBV replication by tetracycline removal for D0 and D10. A representative assay is shown from three independent experiments. (c) Sequential ChIP assays of HepAD38 cells −/+ HBV replication by tetracycline removal for D0 and D10, using BRG1 antibody followed by tandem IP with STAT3 or OCT4 antibody. Results are from three independent experiments. Data were quantified as % of input.
In ESCs the chromatin remodeling BRG1/BAF complex occupies the SALL4 promoter11. Accordingly, we examined whether BRG1 has a role in SALL4 re-expression in HBV infected, differentiated hepatocytes. Accordingly, we performed ChIP assays with BRG1 antibody, representing BAF complex, as a function of HBV replication. Interestingly, BRG1 exhibited increased occupancy of the same SALL4 regulatory region downstream of the TSS that also associated with OCT4 and STAT3, on D10 of HBV replication (Fig. 6a). Reciprocal co-immunoprecipitations using lysates from HepAD38 cells demonstrated that BRG1 interacts with both OCT4 and STAT3, irrespective of HBV replication (Fig. 6b).
To examine whether STAT3, OCT4 and BRG1 are involved in regulation of SALL4 transcription following DNA demethylation of CpG sites downstream of SALL4 TSS, we performed sequential ChIP assays as a function of HBV replication (Fig. 6c). ChIP assays with BRG1 antibody, followed by tandem immunoprecipitation with OCT4 or STAT3 antibody showed enhanced co-occupancy of the SALL4 regulatory region downstream from the TSS, on D10 of HBV replication. These results support that following CpG site demethylation of the SALL4 regulatory region downstream from TSS, OCT4 and STAT3 bind to the respective demethylated binding sites and assemble a complex with BRG1, enabling remodeling of chromatin and SALL4 transcription.
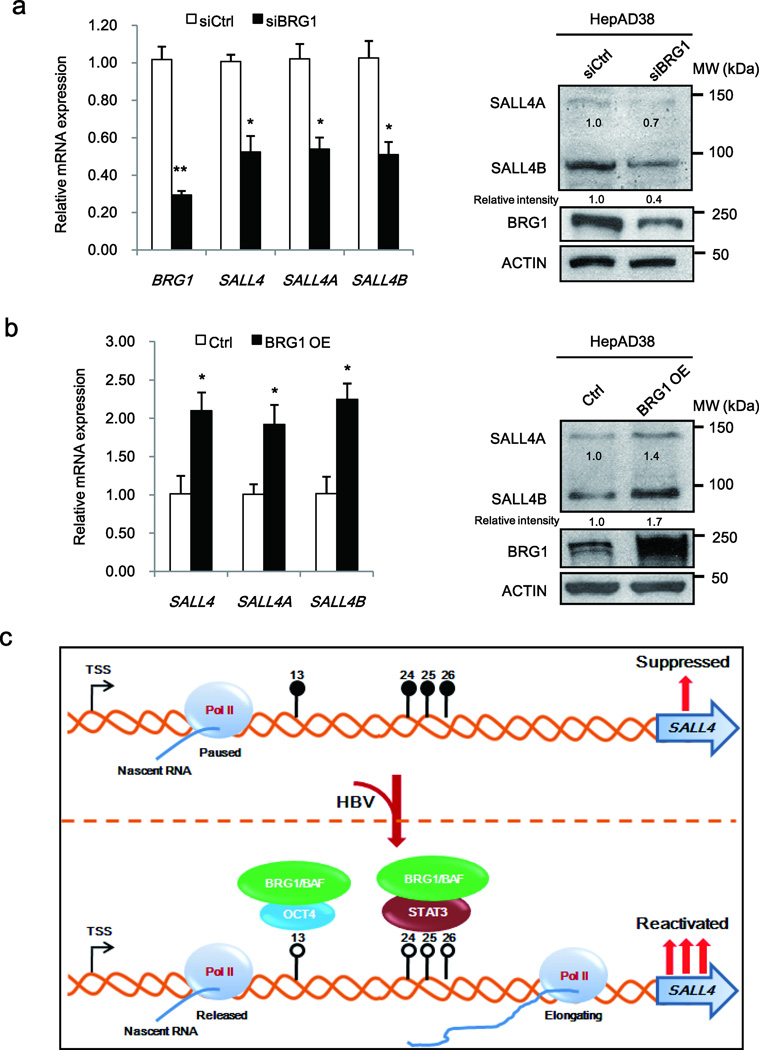
To test this hypothesis, we investigated the functional role of BRG1 in SALL4 transcription. We knocked-down BRG1 on D8 of HBV replication in HepAD38 cells. Indeed, BRG1 knockdown reduced mRNA and protein level of both splicing forms of SALL4, SALL4A and SALLB (Fig. 7a). Conversely, BRG1 overexpression enhanced expression of both SALL4 splicing variants (Fig. 7b). A diagram depicting our working model is shown in Fig. 7c.
Figure 7. BRG1/BAF complex is involved in SALL4 re-expression in HBV replicating hepatocytes.
(a, b) PCR quantification of SALL4 mRNA (Left panel), and immunoblots of SALL4 (Right panel), following transfection of BRG1 siRNA (siBRG1) (a), and BRG1 plasmid (b), in HepAD38 cells grown with HBV replication by tetracycline removal for 10 days. Results are from three independent RNA isolations performed in identical triplicates. Error bars represent S.D. *P< 0.05 and **P< 0.01. BRG1 knockdown and BGR1 overexpression (OE) were confirmed by PCR and immunoblots. (c) A proposed schematic model depicting epigenetic mechanism by which SALL4 is re-expressed during HBV replication.
A positive correlation between SALL4 expression and hepatic cancer stem cell (hCSC) markers in HBV-induced HCCs
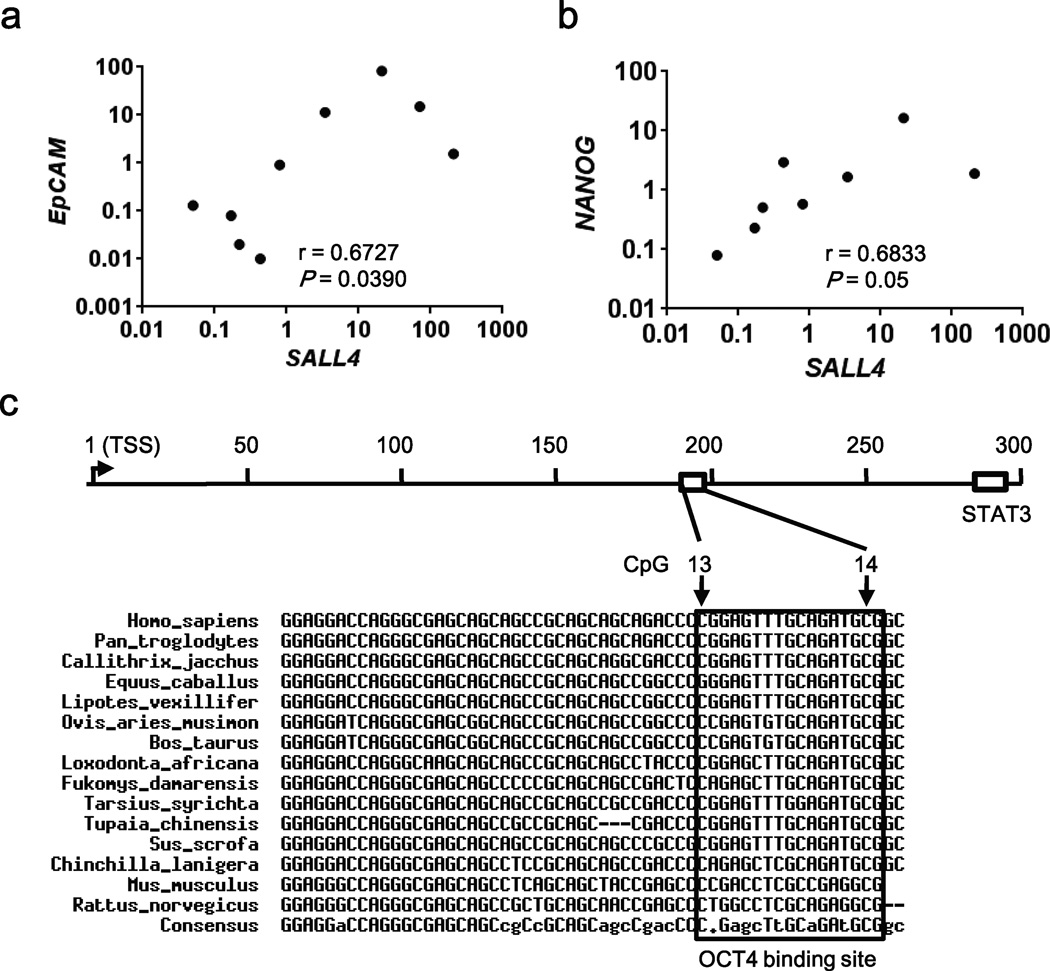
A recent study demonstrated that in mouse embryonic fibroblasts (MEFs), ectopic expression of Sall4, Nanog, Esrrb, and Lin28 (SNEL) generated induced pluripotent stem cells (iPSCs) more efficiently than other factor combinations, including OSKM (Oct4, Sox2, Klf4, and Myc)5. Considering the clinical significance of SALL4 in liver cancer27, 37, 38, and the potential role of SALL4 in prognosis and therapeutics23, we analyzed correlation of SALL4 expression in HBV-related HCCs with expression of NANOG, one of the SNEL factors, and of hepatic cancer stem cell marker EpCAM (Fig. 8). Indeed, a statistically significant correlation was quantified between SALL4 and EpCAM expression in HBV-related HCCs (Fig. 8a), as reported earlier for HCCs. More importantly, a statistically significant correlation also exists between SALL4 and NANOG expression (Fig. 8b), supporting a link of SALL4 to pluripotency. Lastly, sequence analysis shows that the CpG island and OCT4 binding site downstream of SALL4 TSS is conserved across species (Fig. 8c), supporting the essential function of these elements in SALL4 expression.
Figure 8. SALL4 expression correlates with expression of EpCAM and NANOG in HBV-related HCCs.
Scatter plot analysis of SALL4 mRNA expression positively correlated with those of (a) EpCAM and (b) NANOG mRNAs expressed in HBV-related HCCs, quantified by Spearman’s correlation coefficient (r). (c) Sequence analysis of the regulatory region downstream of SALL4 TSS across species. The conserved sequences, CpG sites and OCT4 binding site are shown.
DISCUSSION
It is widely accepted that hepatocarcinogenesis is a long-term process associated with accumulation of genetic and epigenetic alterations, resulting in activation of oncogenes and/or loss of tumor suppressors2. Recent studies have shown deregulated expression of SALL4 in HCC27, 37, 38 and association with poor prognosis37, 38. Although SALL4 transcriptional regulation has been studied in the context of ESCs34, how SALL4 re-expression occurs during hepatocarcinogenesis is not well understood.
In this study, we demonstrate that HBV-related HCCs, characterized by elevated SALL4 expression, display pronounced hypomethylation of the SALL4 regulatory region downstream from the TSS (Figs. 1 and 4). Importantly, the amount of DNA methylation at the SALL4 regulatory region differed between T and PT tissue (Figs. 1 and 4), suggesting changes in DNA methylation of this regulatory region influence levels of SALL4 transcription. We also observed DNA demethylation of this SALL4 regulatory region in the context of HBV infection and replication in vitro (Figs. 2 and 3). Furthermore, employing a methylation-specific assay10 we observed DNA demethylation of these CpG sites in HBV- and HCV-related HCCs, as well as in liver cancer cell lines37 that overexpress SALL4 (Fig. 4). Since there is no report about DNA methylation regulating SALL4 expression in tumors, we retrieved array-based DNA methylation platform data (Illumina HumanMethylation450K BeadChip) from TCGA (The Cancer Genome Atlas, http://cancergenome.nih.gov/) (data not shown). Nineteen probes are available for the genomic region of SALL4, five of which are located in CpG islands. However, all of these CpG sites reside in the gene body of SALL4 and exhibit no DNA methylation differences between T vs. PT tissue. Since DNA methylation in the vicinity of the TSS vs. the gene body13 exerts different effects on transcription, we examined DNA methylation of a CpG island located downstream of the SALL4 TSS (Fig. 1).
Importantly, our findings link hypomethylation of CpG sites in SALL4 regulatory region to enhanced SALL4 transcription. In HBV replicating cells, we quantified increased association of RNA polymerase II with SALL4 chromatin and SALL4 mRNA, employing RNA polymerase II ChIP and RIP assays, respectively (Fig. 5a and b). Our studies also suggest that DNA methylation at CpG sites downstream of the SALL4 TSS result in RNA polymerase II pausing (Fig. 5a). Pausing of RNA polymerase II during early elongation is a widespread regulatory mechanism1. However, further studies are needed to determine whether and how DNA demethylation of specific CpG sites directly regulates RNA polymerase II pausing and SALL4 transcription.
We also show that the mechanism of enhanced SALL4 transcription due to hypomethylation of specific CpG sites involves transcription factors OCT4 and STAT3. In HBV replicating cells, ChIP assays revealed enhanced STAT3 and OCT4 binding to hypomethylated sites located downstream of the SALL4 TSS (Fig. 6). Since STAT3 and OCT4 interact with embryonic stem cell BAF (esBAF) complex and activate transcription of their target genes12, we reasoned that STAT3 and OCT4 may interact with BAF complex in SALL4 regulatory region. Indeed, our co-immunoprecipitation results and more importantly our sequential/tandem ChIP assays with BRG1 and STAT3 or OCT4 antibodies support this hypothesis (Fig. 6c). Consistent with previous genome-wide promoter analysis of BRG1 occupancy of the SALL4 regulatory region in ESCs15, our results show that BRG1 recruitment by STAT3 and OCT4 binding at sites downstream of the SALL4 TSS enable its re-expression in HBV replicating, differentiated hepatocytes (Fig. 7a and b).
OCT4, along with NANOG and SOX2, is expressed exclusively in ESCs28 and is a target of repression by the chromatin modifying polycomb repressive complex 2 (PRC2)30. Our recent studies have identified a molecular mechanism that leads to loss of PRC2 function initiated by the HBV X protein39. Moreover, our new findings show that in the context of loss of PRC2 function, in a subpopulation of HBV replicating cells, there is enhanced expression of pluripotency transcription factors OCT4, NANOG and SOX221, 40, thus identifying cell intrinsic changes occurring in HBV replicating cells that likely contribute to SALL4 re-expression. Indeed, we demonstrate that a positive correlation exists between SALL4, EpCAM and NANOG in HBV-related HCCs, as observed by others29, 38, and that the OCT4 binding site and its associated CpG sites are evolutionarily conserved (Fig. 8). Whether the same cell intrinsic changes involving PRC2 downregulation occur in HCCs of other etiologies, e.g., HCV-associated HCCs, enabling SALL4 expression remain to be determined.
MATERIALS AND METHODS
Tissue samples and cell lines
Paired liver tissues from chronic HBV and HCV patients with HCC, tumor and peritumor were obtained from the French National Biological Resources Center following approved consent from the French Liver Tumor Network Scientific Committee. The HepG2hNTCP 24 and HepAD3817 cell lines that support HBV infection and replication, respectively, were grown as described. Secreted hepatitis B e antigen (HBeAg) was measured by HBeAg ELISA kit (AUTOBIO DIAGNOSTICS CO., Zhengzhou, China). Treatment of indicated cell lines with 10µM 5-Aza-2’-Deoxycytidine (Cayman Chemical, Ann Arbor, MI) was for 48h.
Sodium bisulfite sequencing PCR (BSP)
BSP was performed as previously described7. Human SALL4 CpG island was predicted and bisulfIte sequencing primers were designed by Methprimer18 and listed in Supplementary Table 1. Primers for methylation specific PCR are listed in supplementary Table 1. Equal amount of genomic DNA was amplified under identical amplification conditions.
Chromatin immunoprecipitation (ChIP) assays, immunoprecipitations and immunoblotting
These analyses were carried out as previously described7. The following antibodies were used in this study: SALL4 antibody (Abcam, Cambridge, MA); HBc antibody (Dako, Carpinteria, CA); RNA polymerase II antibody (Santa cruz biotechnology, Dallas, Texas); BRG1 antibody (Abcam, Cambridge, MA); STAT3 antibody (Santa Cruz biotechnology, Dallas, Texas); OCT4 antibody (Abcam, Cambridge, MA); H3K27me3 antibody (Abcam, Cambridge, MA); Histone 3 antibody (Active Motif, Carlsbad, CA). ChIP primer sequences listed in Supplementary Table 1.
RNA immunoprecipitation (RIP)
We followed the standard protocol described previously31. RNA polymerase II antibody (Santa Cruz biotechnology, Dallas, Texas) was used for IP and iScript cDNA Synthesis Kit (Bio-Rad, Hercules, California) was employed for reverse transcription of RNA. RIP primer sequences are available in Supplementary Table 1.
Transient transfections
Transient transfections in HepAD38 cells were performed employing Lipofectamine 2000 (Invitrogen, Grand Island, NY) as described by manufacturer with 10µg of BRG1 mammalian expression plasmid8 (Addgene, Cambridge, MA). BRG1 siRNA and scrambled control siRNA (Invitrogen, Grand Island, NY) were transfected using Lipofectamine RNAi MAX (Invitrogen, Grand Island, NY).
Reverse transcription and quantitative real-time PCR (QPCR)
RNA isolation, reverse transcription and real-time PCR were performed and analyzed as described7. PCR primer sequences are listed in Supplementary Table 1.
Statistical analyses
Statistical significance was assessed by Student’s t-test. Data were expressed as mean ± standard deviation (SD) and *P< 0.05 and **P< 0.01 were considered significant.
Supplementary Material
Acknowledgments
The authors thank the French National Biological Resources Centre for frozen human liver tissues, obtained following approved consent from the French Liver Tumor Network Scientific Committee. The French Liver Tumor Network is funded by the Institut National de la Santé et de la Recherche Médicale (INSERM) and the Agence Nationale de la Recherche (ANR). The authors also thank the Biological Resources Center of Centre Léon Bérard for normal liver tissues obtained following approved consent and ministerial agreement. Financial support: This work was supported by NIH grant DK044533 to OA, and French grants PAIR-CHC 2009 (contract #2009-143, project ENELIVI) from Institute National du Cancer (INCa) to PM. Shared Resources (flow cytometry and DNA sequencing) are supported by NIH grant P30CA023168 to Purdue Center for Cancer Research.
Footnotes
CONFLICT OF INTEREST: The authors who have taken part in this study declared that they do not have anything to disclose regarding funding or conflict of interest with respect to this manuscript.
REFERENCES
- 1.Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nature reviews Genetics. 2012;13:720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aravalli RN, Steer CJ, Cressman EN. Molecular mechanisms of hepatocellular carcinoma. Hepatology. 2008;48:2047–2063. doi: 10.1002/hep.22580. [DOI] [PubMed] [Google Scholar]
- 3.Bard JD, Gelebart P, Amin HM, Young LC, Ma Y, Lai R. Signal transducer and activator of transcription 3 is a transcriptional factor regulating the gene expression of SALL4. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2009;23:1405–1414. doi: 10.1096/fj.08-117721. [DOI] [PubMed] [Google Scholar]
- 4.Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buganim Y, Markoulaki S, van Wietmarschen N, Hoke H, Wu T, Ganz K, et al. The Developmental Potential of iPSCs Is Greatly Influenced by Reprogramming Factor Selection. Cell stem cell. 2014;15:295–309. doi: 10.1016/j.stem.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi JK, Bae JB, Lyu J, Kim TY, Kim YJ. Nucleosome deposition and DNA methylation at coding region boundaries. Genome Biol. 2009;10:R89. doi: 10.1186/gb-2009-10-9-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fan H, Zhang H, Pascuzzi PE, Andrisani O. Hepatitis B virus X protein induces EpCAM expression via active DNA demethylation directed by RelA in complex with EZH2 and TET2. Oncogene. 2016;35:715–726. doi: 10.1038/onc.2015.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gong F, Chiu LY, Cox B, Aymard F, Clouaire T, Leung JW, et al. Screen identifies bromodomain protein ZMYND8 in chromatin recognition of transcription-associated DNA damage that promotes homologous recombination. Genes & development. 2015;29:197–211. doi: 10.1101/gad.252189.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He J, Zhang W, Zhou Q, Zhao T, Song Y, Chai L, et al. Low-expression of microRNA-107 inhibits cell apoptosis in glioma by upregulation of SALL4. The international journal of biochemistry & cell biology. 2013;45:1962–1973. doi: 10.1016/j.biocel.2013.06.008. [DOI] [PubMed] [Google Scholar]
- 10.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ho L, Jothi R, Ronan JL, Cui K, Zhao K, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:5187–5191. doi: 10.1073/pnas.0812888106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ho L, Miller EL, Ronan JL, Ho WQ, Jothi R, Crabtree GR. esBAF facilitates pluripotency by conditioning the genome for LIF/STAT3 signalling and by regulating polycomb function. Nat Cell Biol. 2011;13:903–913. doi: 10.1038/ncb2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nature reviews Genetics. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 14.Jung YK, Jang K, Paik SS, Kwon YJ, Kim HJ, Lee KG, et al. Positive immunostaining of Sal-like protein 4 is associated with poor patient survival outcome in the large and undifferentiated Korean hepatocellular carcinoma. Ann Surg Treat Res. 2016;91:23–28. doi: 10.4174/astr.2016.91.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kidder BL, Palmer S, Knott JG. SWI/SNF-Brg1 regulates self-renewal and occupies core pluripotency-related genes in embryonic stem cells. Stem Cells. 2009;27:317–328. doi: 10.1634/stemcells.2008-0710. [DOI] [PubMed] [Google Scholar]
- 16.Kumaki Y, Oda M, Okano M. QUMA: quantification tool for methylation analysis. Nucleic acids research. 2008;36:W170–W175. doi: 10.1093/nar/gkn294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ladner SK, Otto MJ, Barker CS, Zaifert K, Wang GH, Guo JT, et al. Inducible expression of human hepatitis B virus (HBV) in stably transfected hepatoblastoma cells: a novel system for screening potential inhibitors of HBV replication. Antimicrob Agents Chemother. 1997;41:1715–1720. doi: 10.1128/aac.41.8.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 19.Liao HF, Chen WS, Chen YH, Kao TH, Tseng YT, Lee CY, et al. DNMT3L promotes quiescence in postnatal spermatogonial progenitor cells. Development. 2014;141:2402–2413. doi: 10.1242/dev.105130. [DOI] [PubMed] [Google Scholar]
- 20.Liu H, Wang J, Epner EM. Cyclin D1 activation in B-cell malignancy: association with changes in histone acetylation, DNA methylation, and RNA polymerase II binding to both promoter and distal sequences. Blood. 2004;104:2505–2513. doi: 10.1182/blood-2004-02-0483. [DOI] [PubMed] [Google Scholar]
- 21.Mani SK, Zhang H, Diab A, Pascuzzi PE, Lefrancois L, Fares N, et al. EpCAM-Regulated Intramembrane Proteolysis Induces a Cancer Stem Cell-like Gene Signature in Hepatitis B Virus-infected Hepatocytes. Journal of hepatology. 2016 doi: 10.1016/j.jhep.2016.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mansour AA, Gafni O, Weinberger L, Zviran A, Ayyash M, Rais Y, et al. The H3K27 demethylase Utx regulates somatic and germ cell epigenetic reprogramming. Nature. 2012;488:409–413. doi: 10.1038/nature11272. [DOI] [PubMed] [Google Scholar]
- 23.Marquardt JU, Thorgeirsson SS. Sall4 in "stemness"-driven hepatocarcinogenesis. N Engl J Med. 2013;368:2316–2318. doi: 10.1056/NEJMe1303026. [DOI] [PubMed] [Google Scholar]
- 24.Ni Y, Lempp FA, Mehrle S, Nkongolo S, Kaufman C, Falth M, et al. Hepatitis B and D viruses exploit sodium taurocholate co-transporting polypeptide for species-specific entry into hepatocytes. Gastroenterology. 2014;146:1070–1083. doi: 10.1053/j.gastro.2013.12.024. [DOI] [PubMed] [Google Scholar]
- 25.Nishino K, Toyoda M, Yamazaki-Inoue M, Makino H, Fukawatase Y, Chikazawa E, et al. Defining hypo-methylated regions of stem cell-specific promoters in human iPS cells derived from extra-embryonic amnions and lung fibroblasts. PloS one. 2010;5:e13017. doi: 10.1371/journal.pone.0013017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oikawa T, Kamiya A, Kakinuma S, Zeniya M, Nishinakamura R, Tajiri H, et al. Sall4 regulates cell fate decision in fetal hepatic stem/progenitor cells. Gastroenterology. 2009;136:1000–1011. doi: 10.1053/j.gastro.2008.11.018. [DOI] [PubMed] [Google Scholar]
- 27.Oikawa T, Kamiya A, Zeniya M, Chikada H, Hyuck AD, Yamazaki Y, et al. Sal-like protein 4 (SALL4), a stem cell biomarker in liver cancers. Hepatology. 2013;57:1469–1483. doi: 10.1002/hep.26159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pan GJ, Chang ZY, Scholer HR, Pei D. Stem cell pluripotency and transcription factor Oct4. Cell research. 2002;12:321–329. doi: 10.1038/sj.cr.7290134. [DOI] [PubMed] [Google Scholar]
- 29.Park H, Lee H, Seo AN, Cho JY, Choi YR, Yoon YS, et al. SALL4 Expression in Hepatocellular Carcinomas Is Associated with EpCAM-Positivity and a Poor Prognosis. J Pathol Transl Med. 2015;49:373–381. doi: 10.4132/jptm.2015.07.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Selth LA, Gilbert C, Svejstrup JQ. RNA immunoprecipitation to determine RNA-protein associations in vivo. Cold Spring Harbor protocols. 2009;2009 doi: 10.1101/pdb.prot5234. pdb prot5234. [DOI] [PubMed] [Google Scholar]
- 32.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA: a cancer journal for clinicians. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 33.Veloso A, Kirkconnell KS, Magnuson B, Biewen B, Paulsen MT, Wilson TE, et al. Rate of elongation by RNA polymerase II is associated with specific gene features and epigenetic modifications. Genome research. 2014;24:896–905. doi: 10.1101/gr.171405.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang J, Gao C, Chai L, Ma Y. A novel SALL4/OCT4 transcriptional feedback network for pluripotency of embryonic stem cells. PloS one. 2010;5:e10766. doi: 10.1371/journal.pone.0010766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang J, Corsello TR, Ma Y. Stem cell gene SALL4 suppresses transcription through recruitment of DNA methyltransferases. The Journal of biological chemistry. 2012;287:1996–2005. doi: 10.1074/jbc.M111.308734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yin F, Han X, Yao SK, Wang XL, Yang HC. Importance of SALL4 in the development and prognosis of hepatocellular carcinoma. World J Gastroenterol. 2016;22:2837–2843. doi: 10.3748/wjg.v22.i9.2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yong KJ, Gao C, Lim JS, Yan B, Yang H, Dimitrov T, et al. Oncofetal gene SALL4 in aggressive hepatocellular carcinoma. N Engl J Med. 2013;368:2266–2276. doi: 10.1056/NEJMoa1300297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zeng SS, Yamashita T, Kondo M, Nio K, Hayashi T, Hara Y, et al. The transcription factor SALL4 regulates stemness of EpCAM-positive hepatocellular carcinoma. J Hepatol. 2014;60:127–134. doi: 10.1016/j.jhep.2013.08.024. [DOI] [PubMed] [Google Scholar]
- 39.Zhang H, Diab A, Fan H, Mani SK, Hullinger R, Merle P, et al. PLK1 and HOTAIR Accelerate Proteasomal Degradation of SUZ12 and ZNF198 during Hepatitis B Virus-Induced Liver Carcinogenesis. Cancer research. 2015;75:2363–2374. doi: 10.1158/0008-5472.CAN-14-2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang H, Xing Z, Mani SK, Bancel B, Durantel D, Zoulim F, et al. RNA helicase DDX5 regulates PRC2/HOTAIR function in Hepatitis B Virus infection and hepatocarcinogenesis. Hepatology. 2016 doi: 10.1002/hep.28698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang J, Tam WL, Tong GQ, Wu Q, Chan HY, Soh BS, et al. Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1. Nat Cell Biol. 2006;8:1114–1123. doi: 10.1038/ncb1481. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.