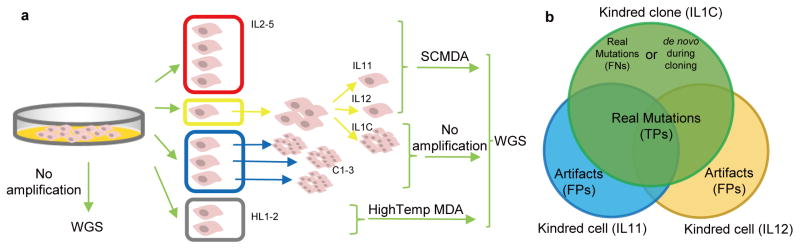
Figure 1. Experimental design for validating SNV identification in SCMDA-amplified single cells.
(a) To allow validation for accurate single cell amplification and variant calling, whole genome sequencing (WGS) was performed on (1) four single cells amplified using SCMDA (red); (2) two cells amplified using SCMDA and their non-amplified kindred clone (yellow); (3) three additional, unamplified clones (blue); and (4) two single cells amplified after high temperature lysis (grey). Cell / clone IDs are included in the Figure. (b) The kindred cells and clone are expected to have identical genotypes, including both germline and somatic SNVs. Candidate SNVs identified in both clone and single cells are true positives (TPs). Those found in neither of the cells but only in the clone are false negatives (FNs). Variants found only in one cell are considered false positives (FPs). See Supplementary Note for details. These are conservative assumptions and do not take into account possible de novo mutations in the kindred clone or single cells arising after their divergence. Of note, such events would increase sensitivity and specificity.