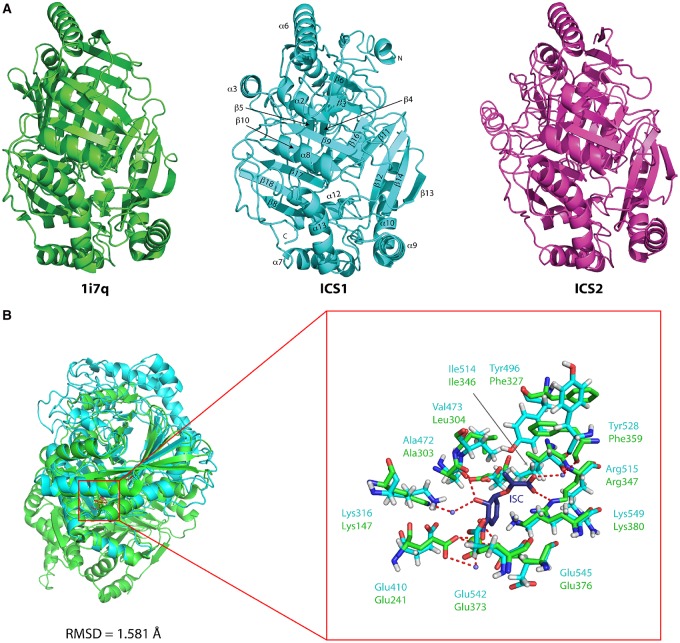
Figure 1. Homology models of Arabidopsis ICS1 and ICS2.
(A) Primary amino acid sequences for the two Arabidopsis ICS isozymes were submitted to the iTASSER homology modelling server [29] for structural alignment with existing proteins structures. The reference structure, S. marcescens anthranilate synthase (TrpE; PDB accession 1I7Q), is shown alongside the models obtained with this structure for ICS1 and ICS2. α-helices and β-sheets are numbered according to the scheme shown in Supplementary Figure S3; some labels are omitted here for clarity. Amino (N) and carboxy (C) termini are labelled accordingly. ICS1 and ICS2 adopt the same overall structure, with identical arrangement of α-helices and β-sheets, as might be expected for two paralogous proteins. Images obtained using MacPyMOL (http://www.pymol.org). (B) Comparison of ICS1 (pale blue) with E. coli EntC (green), showing very similar active sites.