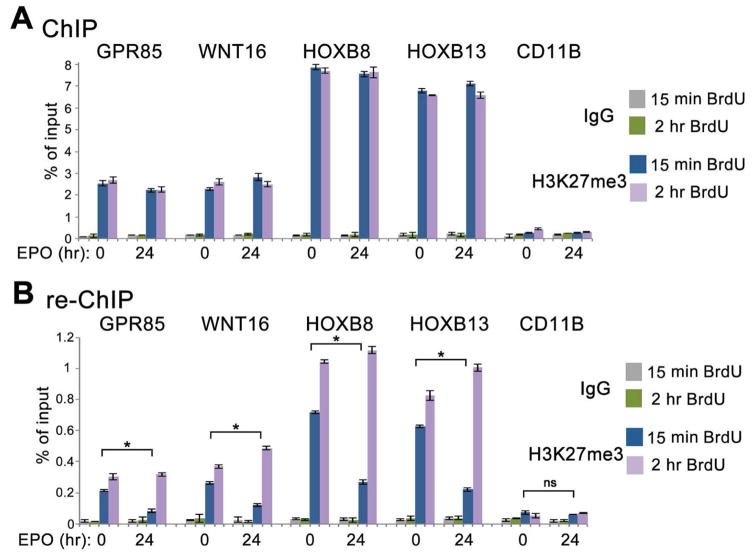
Figure 6. Kinetics of H3K27me3 accumulation at nascent DNA of specific genes during erythroid differentiation of CD34+ HPCs.
(A) ChIP assays with the H3K27me3 antibody. Uninduced and 24 hr EPO-induced CD34+ cells were labeled with BrdU for 15 min or 15 min and chased to 60 min. Chromatin was immunoprecipitated with H3K27me3 and IgG antibodies.
(B) ChIP assays with the BrdU antibody. DNA from the first immunoprecipitation step with H3K27me3 antibody (A) was denatured and immunoprecipitated with the BrdU antibody. Q-PCR was performed at genes repressed before and after EPO induction (GPR85, WNT16, HOXB8, HOXB13) and at a non-repressed control gene (CD11B) (Cui et al., 2009). Percent of input for re-ChIP experiments was calculated using the amount of material eluted from the beads following immunoprecipitation with H3K27me3 as 100% input. Data are represented as mean ± SD of technical replicates of one of the independent experiments. p-values were calculated by a one-way ANOVA with Bonferroni’s post-hoc test. The statistical significance was defined as p<0.05.