Abstract
Motivation
Chloroplasts are organelles found in plants and involved in several important cell processes. Similarly to other compartments in the cell, chloroplasts have an internal structure comprising several sub-compartments, where different proteins are targeted to perform their functions. Given the relation between protein function and localization, the availability of effective computational tools to predict protein sub-organelle localizations is crucial for large-scale functional studies.
Results
In this paper we present SChloro, a novel machine-learning approach to predict protein sub-chloroplastic localization, based on targeting signal detection and membrane protein information. The proposed approach performs multi-label predictions discriminating six chloroplastic sub-compartments that include inner membrane, outer membrane, stroma, thylakoid lumen, plastoglobule and thylakoid membrane. In comparative benchmarks, the proposed method outperforms current state-of-the-art methods in both single- and multi-compartment predictions, with an overall multi-label accuracy of 74%. The results demonstrate the relevance of the approach that is eligible as a good candidate for integration into more general large-scale annotation pipelines of protein subcellular localization.
Availability and Implementation
The method is available as web server at http://schloro.biocomp.unibo.it
1 Introduction
The eukaryotic cell hosts different compartments that play differentiated functional roles into the cell life cycle. Chloroplasts are organelles found in viridiplantae cells and involved in crucial functions including photosynthesis, fatty acid synthesis and immune response. Similarly to other compartments in the cell, such as the nucleus or mitochondria, in-depth experimental studies have identified at least six different chloroplastic sub-compartments in which proteins are targeted to perform different functions (Cooper and Hausman, 2009): the inner membrane, the outer membrane, the stroma, the thylakoid lumen, the plastoglobule and the thylakoid membrane.
Few proteins found in the chloroplast are encoded by the organelle genome whereas the vast majority of them are nuclear encoded, synthesized by cytoplasmic ribosomes and then post-translationally targeted into the chloroplast by means of different mechanisms (Schleiff and Becker, 2010). Generally, targeting signals are present in the precursor protein and are used by the transport machinery to correctly direct the protein to its final destination. Most proteins directed to the stroma or to the envelope carry a single cleavable N-terminal signal, while proteins directed to the thylakoid lumen and membrane are endowed with a bipartite signal, which provides information for the subsequent sorting of the protein from the stroma to the thylakoid. Furthermore, several non-cleavable sequence signals may also be present at any position along the sequence (typically membrane proteins are endowed with this type of signals) (Schleiff and Becker, 2010). In general, the import and sorting machinery is able to recognize these signals and to transport both soluble proteins (directed to the stroma or to the thylakoid lumen) and membrane proteins (directed to the thylakoid membrane or to the envelope) with single or multiple trans-membrane domains to their final working compartment (Schleiff and Becker, 2010).
So far, several computational tools have been developed to predict protein subcellular localization, given the impact of the feature on protein function characterization (Imai and Nakai, 2010).
The vast majority of available computational methods routinely discriminate macro compartments such as nucleus, cytoplasm, organelles and membranes (Emanuelsson et al., 2007; Goldberg et al., 2014; Marcotte et al., 2000; Nakai and Horton, 1999; Nair and Rost, 2005; Savojardo et al., 2015). However, the prediction of more detailed sub-localizations, such as the different sub-chloroplastic compartments, is challenging considering the paucity of detailed experimental annotations in publicly available databases (e.g. UniprotKB). For instance, only half of the currently available chloroplastic proteins with experimental evidence have also a sub-chloroplastic experimental annotation. Nonetheless, there has been a renewed interest in developing computational tools that are able to correctly identify very specific cellular sub-compartments (Kumar et al., 2014; Lin et al., 2013; Wang et al., 2015).
The prediction of sub-chloroplastic localization has been mainly addressed in two ways: (i) single-label approaches, which associate to the query protein a single localization compartment (Du et al., 2009; Hu and Yan, 2012; Shi et al., 2011; Tung et al., 2010) and (ii) multi-label approaches that can predict multiple localizations (Wang et al., 2015).
Generally, single-label methods consider four main chloroplastic sub-compartments: envelope, stroma, thylakoid lumen and thylakoid membrane. All of them are based on similar features extracted from protein sequence, which are then processed by different algorithms to perform the final prediction. SubChlo (Du et al., 2009), one the first released methods, is based on a variant of the k-nearest neighbor classifier and Chou’s pseudo amino-acid composition (PseAAC) (Chou, 2001). In ChloroRF (Tung et al., 2010), a random forest classifier is fed with a protein encoding based on physicochemical properties extracted from the AAindex (Kawashima et al., 2008). SubIdent (Shi et al., 2011), which can also predict sub-mitochondrial localizations, performs predictions using SVMs and an alternative formulation of the PseAAC based on discrete wavelet transform. Finally, BS-KNN (Hu and Yan, 2012) is based on bit-score k-nearest neighbor and standard amino acid composition.
The only available multi-label method is MultiP-SChlo (Wang et al., 2015). It extends the set of possible compartments in which a protein can be found, by including plastoglobules, lipoprotein particles present in all plastids. Then, using an algorithm based on multi-stage SVMs and PseAAC, the method performs multi-label predictions. MultiP-SChlo scores with an overall accuracy of 56% on a benchmark of a multi-label dataset introduced in the same study (Wang et al., 2015).
In this paper we present SChloro, a novel machine-learning method to improve the prediction of protein sub-chloroplastic localization. The basic idea of our approach is to exploit the recognition of high-level topological and sorting features to improve the accuracy of the prediction of sub-chloroplastic localization. We adopt a two-stage prediction algorithm: first, we identify into the query protein, chloroplastic and/or thylakoid sorting signals and second, we determine possible membrane interactions (suggesting membrane-related localizations). In the final step, these predicted features are integrated with global protein features to predict the final sub-chloroplastic localization, in a multi-label fashion. Differently from any previous approach, our method is able to provide predictions to six distinct compartments: inner membrane, outer membrane, stroma, plastoglobule, thylakoid lumen and thylakoid membrane. When compared to other state-of-the-art approaches, SChloro is able to significantly improve the prediction performance, scoring with a 74% overall multi-label accuracy. The method is available as web server at http://schloro.biocomp.unibo.it.
2 Methods
2.1 Datasets
In this study, three different datasets were used to evaluate the performance of our method and to compare it with previously developed approaches.
2.1.1 The SCEXP2016 dataset
The first dataset, referred to as SCEXP2016, was specifically compiled for this study and collects updated experimental data extracted from UniprotKB/SwissProt release 2016_01 (The UniProt Consortium, 2014). In order to retain only high-quality data, the following procedure was adopted. Firstly, all chloroplastic proteins with experimentally annotated sub-cellular localization were extracted from UniprotKB/SwissProt. Only proteins with evidence at the protein level and longer than 50 residues were selected. From this initial set, to obtain very clean data, we filtered-out proteins that were annotated with additional localizations outside the chloroplast and retained only those with experimental annotation in at least one of the following six chloroplastic sub-compartments: inner membrane, outer membrane, stroma, plastoglobule, thylakoid lumen and thylakoid membrane. With this procedure, we ended up with 367 protein sequences, 309 of which are nuclear encoded whereas 26 are encoded by the chloroplastic genome (we decided to retain these proteins given the small number). Twenty-three out of 367 proteins are annotated with multiple chloroplastic sub-compartments (22 found in two compartments and 1 in three compartments).
The distribution of proteins into the six different chloroplastic sub-compartments is summarized in Table 1. Furthermore, in Table 2 we also list the statistics of targeting signal and membrane interaction annotations (which will be used to train/test specific classifiers, as described in Section 2.4). It is worth to point out that, as detailed above, experimental evidence has been checked only for the primary annotation of proteins into subcellular compartments. In contrast, secondary protein annotation concerning targeting signals and membrane interaction were all retained and used as they were annotated for the selected proteins. As a consequence, these secondary annotations could be partially incomplete
Table 1.
Distribution of proteins in SCEXP2016 into the six different chloroplastic sub-compartments
| Compartment | Number of proteins |
|---|---|
| Inner membrane | 47 |
| Outer membrane | 24 |
| Stroma | 119 |
| Plastoglobule | 32 |
| Thylakoid lumen | 37 |
| Thylakoid membrane | 131 |
Table 2.
Distribution of annotated targeting and membrane features of SCEXP2016 proteins
| Feature | Number of annotated proteins |
|---|---|
| Chloroplastic targeting | 317 |
| Thylakoid targeting | 60 |
| Single-pass membrane | 34 |
| Multi-pass membrane | 62 |
| Peripheral membrane | 41 |
In order to avoid any training/test bias, cross-validation sets were built by confining any possible local sequence homology into the same validation set. To achieve this, we firstly searched each protein sequence against the whole dataset using the psi-blast program with e-value threshold set to 1e-3. Sequence clusters were then built using the psi-blast output. In particular, two sequences fell into the same cluster if psi-blast detected at least one hit between them (no identity threshold was set for cluster generation). These clusters were finally used to compile 10 cross-validation sets for method evaluation.
2.1.2 The MSchlor578 dataset
The second dataset adopted in this study is the MSchlo578 dataset, previously released by Wang et al. (2015). This dataset contains 578 multi-compartment proteins distributed into the five following sub-chloroplastic localizations (in parenthesis the number of proteins): envelope (199), stroma (105), thylakoid lumen (34), thylakoid membrane (233) and plastoglobule (30). Twenty-two proteins are annotated with multiple sub-compartments (21 into two different compartments and 1 in three compartments). We used the MSchlo578 dataset to compare our method with the state-of-the-art method MultiP-Schlo (Wang et al., 2015).
2.1.3 The S60 dataset
Finally, a third dataset, referred to as S60 and introduced by Du et al. (2009), was used to compare our method with other methods in the single-label setting. The 262 proteins in this dataset are distributed among 4 different classes: envelope (40), stroma (49), lumen (44) and thylakoid membrane (129). No multiple annotations are reported for these proteins.
2.2 Sorting signals to chloroplast and its sub-compartments
Nuclear encoded chloroplastic proteins are targeted toward the organelle by means of biological pathways involving the molecular recognition of specific sorting signals (Schleiff and Becker, 2010). At a higher level, precursor proteins synthesized by cytoplasmic ribosomes, are endowed with the well-known transit peptide, a variable-length stretch of sequence located at the N-terminus of the nascent protein (Bruce, 2001; Patron and Waller, 2007; Schleiff and Becker, 2010). Once the protein reaches its destination into the chloroplast (typically the stroma), the transit peptide is cleaved by specific proteins. Some chloroplastic proteins of the thylakoid lumen and membranes are endowed with an additional signal located immediately after the transit peptide. This thylakoid transit peptide is used for the subsequent protein sorting from the stroma to the thylakoid (Bruce, 2001; Schleiff and Becker, 2010).
In addition, a subset of nuclear-encoded chloroplastic proteins was found as not having the classic transit peptide. These proteins are mainly outer-membrane proteins (and also inner-membrane and inter-membrane space proteins, although to a lesser extent) with alpha helical membrane anchors, which also carry targeting information (Schleiff and Klösgen, 2001; Soll, 2002).
In this paper we try to exploit the knowledge about these mechanisms by defining signal-specific detectors and integrating them into our localization prediction system (see Section 2.4 for details).
2.3 Membrane interaction
The structure of the chloroplasts comprises three different membrane systems: the inner and outer membranes and the thylakoid membrane system. The inner and outer membranes form the chloroplast envelope, which borders the stroma, and separates it from the cytoplasm. Inside the stroma, it is found the thylakoid, an additional membrane-bounded structure. The thylakoid membrane separates the stroma from the lumen. Several membrane proteins with diverse topologies can be found as either directly or indirectly interacting with the three membrane systems. According to the type of interaction, three major classes can be distinguished:
Integral single-pass membrane proteins, which spans the membrane with a single trans-membrane domain.
Integral multi-pass membrane proteins, which spans the membrane with multiple trans-membrane domains.
Peripheral membrane proteins, which do not span the membrane and interact with it through different mechanisms including lipid anchoring, direct interaction with the phospholipid bilayer through specific domains or indirect interaction though integral membrane proteins.
From the point of view of protein sub-chloroplastic localization prediction, knowing whether a protein interacts or not with a membrane may directly restrict the number of possible compartments it may be found in. Furthermore, the precise knowledge of the interaction type (single-, multi-pass or peripheral) may give some additional insight about the final destination of the protein. In this paper, we exploited these considerations by integrating membrane-interaction specific classifiers into our localization prediction system (see Section 2.4 for details).
2.4 Overview of the prediction method
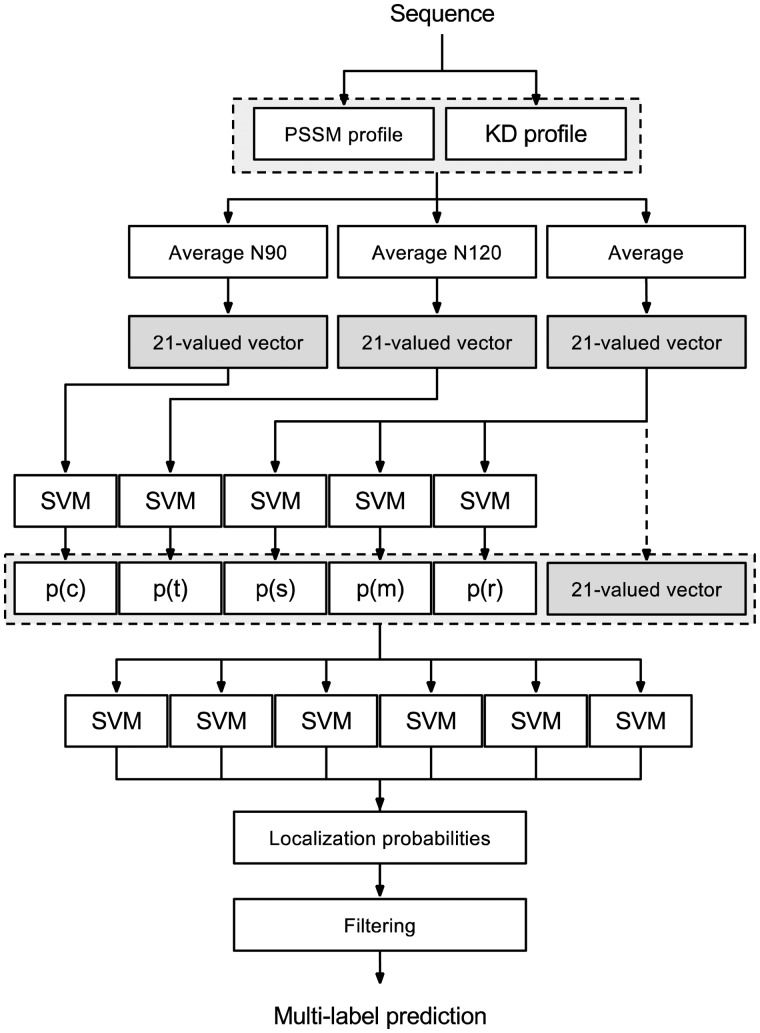
The proposed multi-label prediction system, depicted in Figure 1, consists of two layers of Support Vector Machines (SVMs). Classifiers of the first layer are devised to predict the occurrence probabilities of chloroplast and/or thylakoid sorting signals as well as the probabilities for the protein to be in one of three possible interaction states with a membrane (single-, multi-pass trans-membrane or peripheral membrane protein). Therefore, five different classifiers were defined: two for the sorting signals and three for the membrane interaction. Each classifier was trained using available experimental evidence and slightly different input features optimized for the specific prediction task. In particular the following input features are used here:
Fig. 1.
Overview of the SChloro system architecture
The average composition of the Position Specific Scoring Matrix (PSSM) as computed from the multiple sequence alignment obtained using the psi-blast program (Altschul et al., 1997) to search the query sequence against the UniprotKB/SwissProt database (The UniProt Consortium, 2014). Raw PSSM values are rescaled before averaging into the range [0,1] using a standard logistic function 1/((1+e^(-x))). In this way, the average PSSM consists of a 20-valued vector with elements ranging between 0 and 1.
The average hydrophobicity computed along the protein sequence using the Kyte-Doolittle scale (Kyte and Doolittle, 1982). Hydrophobicity values are firstly linearly rescaled before averaging into the range [0,1] so that the highest and the lowest values, namely 4.5 and -4.5 for isoleucine and arginine, map to 0 and 1, respectively. Hence, the average hydrophobicity feature consists of a single real value between 0 and 1.
For the chloroplast and thylakoid targeting classifiers, considering that the two targeting signals are located at the N-terminus of the protein, the first 90 and 120 residues were used to compute the average values, respectively. In contrast, the entire protein sequence has been used for the three membrane interaction classifiers. Altogether, the first layer outputs are collected into a 5-valued vector defined as follows:
| (1) |
where the first two values are, respectively, the probabilities of having a chloroplastic-targeting signal (p(c)) and thylakoid-targeting signal (p(t)), while the last three values are the probabilities for the protein to be, respectively, a single-pass (p(s)), a multi-pass (p(m)) and a peripheral (p(r)) membrane protein.
The second layer of SVM classifiers computes the membership probability for the query protein to be located into one or more sub-chloroplastic compartments. The number of independent SVMs of the second layer is determined by the number of the predicted localization classes. The final version of SChloro is able to predict six different sub-chloroplastic localizations. As a consequence, one separate SVM classifier was defined for each one of the six compartments. Each second-layer classifier was trained using a 26-valued feature vector consisting of: (i) the 5-valued vector as defined in Equation 1 and (ii) the average PSSM and hydrophobicity both computed on the entire protein sequence.
Finally, the individual SVM output probabilities are integrated into the final multi-label prediction of the target sequence. In particular, the protein is predicted as belonging to one localization class if the corresponding SVM probability output is greater or equal to 0.5. The multi-label prediction is simply obtained by the union of all the individual localization predictions.
Adopting this two-layered architecture allows a better exploitation of different basic features that are computed over different portions of the sequence. By this, an intermediate representation of the protein in terms of presence/absence of sorting signals as well as interaction with the membrane, is computed.
2.5 Model selection and implementation
The method evaluations are carried-out using either a 10-fold cross-validation procedure (to train/test our method on the SCEXP2016 dataset), or by adopting a jackknife test (to compare with other methods in literature on the MSchlor578 and S60 datasets). Regardless of the performed actual evaluation setting, the benchmark procedure needs to be carefully tuned to deal with the specific structure of our prediction system that comprises two cascading levels of classifiers.
To achieve this, we applied the following procedure. First of all, for each cross-validation or jackknife run, a fraction of the training set was extracted and used as a validation set. This set was used to adjust hyper-parameters as well as to identify the optimal input feature encoding for both first- and second-layer classifiers. Once selected, these hyper-parameters were frozen and used to predict the remaining testing data.
SVM classifiers were implemented using the standard libsvm software package (Chang et al., 2011). Each classifier is based on a non-linear Radial Basis Function (RBF) kernel and is trained/tested to provide probabilistic outputs using the standard model implemented by the software library.
Concerning the cascading structure, optimal first-layer classifiers (found through validation sets) were used to generate both training/testing data for second-layer classifiers. In this way, SVMs of the second-layer were trained/tested on predicted values and this allowed evaluating the entire pipeline taking into account the potential error propagation between the two layers.
2.6 Scoring measures
For sake of comparison with different methods available in literature, our system was evaluated using either multi-label or single-label scoring measures. More formally, let and be the set of observed and predicted labels (compartments) for the ith protein, and let be the total number of proteins in the dataset. To score the prediction performance in the multi-label setting, we adopted the following scoring indexes (Wang et al., 2015):
- The multi-label Accuracy (mlACC), defined as:
(2) - The multi-label Recall (mlREC), defined as:
(3) - The multi-label Precision (mlPRE), defined as:
(4) - The multi-label F1 (mlF1), defined as:
(5) - The overall multi-label accuracy (ACCml), defined as:
(6)
where is an indicator function that equals to 1 if the two sets are identical, 0 otherwise.
To score the prediction performance in the single-label setting we used the following scoring indexes (Du et al., 2009):
- The single-label accuracy of label (ACCsl(l)), defined as:
(7) - The overall single-label accuracy (ACCsl), defined as:
(8)
where TPl and FNl are true positive and false negatives for the label l, respectively, and m is the number of different labels.
Finally, each classifier in the first layer was scored using the Matthews Correlation Coefficient (MCC) and the Area Under the ROC Curve (AUC), defined as:
| (9) |
| (10) |
where TP, TN, FP and FN are true positives, true negatives, false positives and false negatives, respectively, Sen(t) and FPR(t) are standard sensitivity and false positive rate values, respectively, computed fixing the prediction threshold to t (the outputs of each classifiers are probabilities).
3 Results
3.1 Single- and multi-label performance of SChloro on the SCEXP2016 dataset
Table 3 lists the 10-fold cross-validation results obtained using different input features and evaluated on the SCEXP2016 dataset. Both single- and multi-label scoring indexes are reported. The baseline predictor (first row in Table 3) does not include information about targeting signals and membrane interaction and it was trained/tested using the basic feature encoding (consisting of average PSSM and hydrophobicity computed on the entire protein sequence. In this case, only the second layer of the SVM system is used).
Table 3.
Single- and multi-label performance with different combinations of input features on the SCEXP2016 dataset by adopting a 10-fold cross-validation procedure
| Multi-label prediction |
Single-label prediction |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Input features | ACCml | mlACC | mlPRE | mlREC | mlF1 | ACCsl(I) | ACCsl(O) | ACCsl(S) | ACCsl(L) | ACCsl(M) | ACCsl(P) | ACCsl |
| Basic | 0.48 | 0.56 | 0.56 | 0.61 | 0.59 | 0.33 | 0.26 | 0.60 | 0.61 | 0.57 | 0.48 | 0.58 |
| Basic+target (predicted) | 0.62 | 0.75 | 0.76 | 0.90 | 0.83 | 0.62 | 0.57 | 0.89 | 0.89 | 0.82 | 0.80 | 0.89 |
| Basic+mem (predicted) | 0.60 | 0.74 | 0.75 | 0.89 | 0.82 | 0.67 | 0.65 | 0.84 | 0.85 | 0.83 | 0.75 | 0.85 |
| Basic+target+mem (predicted) | 0.63 | 0.77 | 0.79 | 0.93 | 0.85 | 0.66 | 0.63 | 0.90 | 0.89 | 0.84 | 0.81 | 0.91 |
| Basic+target+mem (observed) | 0.73 | 0.82 | 0.84 | 0.93 | 0.89 | 0.81 | 0.75 | 0.97 | 0.97 | 0.92 | 0.86 | 0.96 |
Basic = PSSM + Hydrophobicty; target = [p(c),p(t)]; mem = [p(s),p(m),p(r)]. Scoring indexes are defined as in Section 2.3. In single-label scoring indexes, I, O, S, L, M and P stand for inner membrane, outer membrane, stroma, thylakoid lumen, thylakoid membrane and plastoglobule, respectively.
The individual contributions of the two feature types (targeting signals and membrane interactions) are reported in rows 2 and 3 of Table 3, respectively. As expected, the inclusion of the targeting feature has a major impact in predicting targeting-related localizations (i.e. stroma, lumen and thylakoid membrane). On the contrary, membrane interaction features are more effective in predicting membrane-related localizations (in particular inner and outer membranes).
When predicted probabilities of targeting signals and membrane interaction are both included, the prediction performance becomes more balanced and generally improves (compare rows 1–4, in Table 3). In particular, we observe a general improvement in performance, with ACCml increasing up to 0.63 and ACCsl up to 0.91. Furthermore, also individual single-label accuracies improve, suggesting a general positive contribution of the five predicted features.
For sake of comparison, we also report results obtained when the real information about targeting signals and membrane interaction is included in the second step of the procedure (i.e. in both training and testing, predicted probabilities are replaced by binary features derived from the true annotation of each protein). The reported performance scores represent the maximum theoretical accuracy that can be achieved on this dataset assuming a perfect targeting and membrane interaction prediction. This theoretical predictor achieves very high overall accuracies (ACCml = 0.73 and ACCsl = 0.96), suggesting that the proposed approach builds on top of sound bases and that the prediction performance might be further improved by providing more accurate first-level feature predictors.
Finally, for sake of completeness, in Table 4 we also report the performance of individual first-layer classifiers devised to predict sorting signals and membrane interaction. Considering the results and the inherent difficulty of each prediction task, it appears that the effectiveness of individual predictors is strongly affected by the corresponding abundance of the annotated data in the dataset (compare Tables 2 and 4).
Table 4.
10-Fold cross-validation performance of SChloro classifiers for targeting signals and membrane interactions
| Classifier | AUC | MCC |
|---|---|---|
| Chloroplast targeting | 0.85 | 0.76 |
| Thylakoid targeting | 0.94 | 0.70 |
| Single-pass membrane | 0.82 | 0.47 |
| Multi-pass membrane | 0.93 | 0.67 |
| Peripheral membrane | 0.80 | 0.37 |
3.2 Comparison with other single- and multi-label methods
In Table 5 we report a comparative benchmark of different methods on the S60 dataset (Du et al., 2009). For sake of comparison, results of SChloro were computed using a jackknife test, while performance scores for other methods were taken from literature (Wang et al., 2015). In particular, we report overall single-label accuracies using the same annotation scheme consisting of four different labels (E = envelope, S = stroma, L = lumen, M = thylakoid membrane), respectively, for our method and for other five different single-label methods available in literature: SubChlo (Du et al., 2009), ChloroRF (Tung et al., 2010), SubIdent (Shi et al., 2011), BS-KNN (Hu and Yan, 2012) and MultiP-Schlo (Wang et al., 2015). The results indicate that SChloro provides in general more balanced predictions compared to others. Other methods tend to over-predict the more abundant labels in the dataset (i.e. thylakoid membrane and stroma), whereas SChloro scores, on average, better on overall accuracy and in all the remaining compartments (e.g. compare accuracy results for the lumen and envelope labels).
Table 5.
Comparison of single-label performance of different methods on the S60 dataset adopting a jackknife test
| Method | ACCsl | ACCsl(E) | ACCsl(S) | ACCsl(L) | ACCsl(M) |
|---|---|---|---|---|---|
| SChloro | 0.90 | 0.93 | 0.96 | 0.98 | 0.89 |
| MultiP-Schloa | 0.89 | 0.73 | 0.96 | 0.61 | 1.0 |
| SubChloa | 0.67 | 0.40 | 0.67 | 0.43 | 0.84 |
| ChloroRFa | 0.67 | 0.48 | 0.57 | 0.39 | 0.88 |
| SubIdenta | 0.89 | 0.80 | 0.86 | 0.64 | 0.98 |
| BS-KNNa | 0.76 | 0.48 | 0.74 | 0.78 | 0.85 |
Scoring indexes are defined in Section 2.3. Labels E, S, L and M stand for envelope, stroma, thylakoid lumen and thylakoid membrane, respectively.
Data taken from Wang et al. (2015)
Finally, multi-label prediction performances are reported in Table 6. Here we compare SChloro with MultiP-Schlo (Wang et al., 2015). In this case, results reported for our method are computed using the same annotation scheme of MultiP-Schlo, including five compartments: envelope, stroma, lumen, thylakoid membrane and plastoglobule. In this benchmark, we obtain a significant improvement. SChloro outperforms MultiP-Schlo in all scoring indexes reported, achieving an improvement of about 12% in overall multi-label accuracy.
Table 6.
Comparison of multi-label performance of MultiP-Schlo and our method on the MSchlo578 dataset
| Method | ACCml | mlACC | mlPRE | mlREC | mlF1 |
|---|---|---|---|---|---|
| SChloro | 0.74 | 0.76 | 0.78 | 0.78 | 0.78 |
| MultiP-Schlo | 0.56 | 0.63 | 0.64 | 0.71 | 0.67 |
Scoring indexes are defined in Section 2.3. The comparison adopts a jackknife test.
4 Conclusion
Assessing the protein sub-cellular localization is an important step toward protein function prediction. The rapid pace at which new proteomes become available through NGS technologies requires the availability of effective computational tools for assessing protein localization and function to fill the gaps of the experimental knowledge.
In this paper we presented SChloro, a novel approach to predict protein sub-chloroplastic localization into six main compartments including inner and outer membranes, stroma, plastoglobule, lumen and thylakoid membrane. Our method is based on the recognition of sequence signals that define target specificity (chloroplast and thylakoid targeting signals) as well as on the prediction of the potential type of interaction with chloroplast membranes (single-pass, multi-pass and peripheral interaction). We show that this information can be profitably incorporated into a two-level SVM-based algorithm to predict both single and multiple protein sub-chloroplastic localizations with high accuracy. In fact, SChloro significantly outperforms the available state-of-the-art methods, both in single and multi-label settings. Furthermore, regardless of the specific dataset and evaluation setting adopted, the performance of SChloro resulted rather stable throughout all the experiments performed, showing that our approach is sufficiently robust and not so sensitive to the specific dataset chosen. This fact makes SChloro a good candidate for the integration into a more comprehensive pipeline for the annotation of sub-cellular localization of protein in plant organisms.
The complete prediction system is available as web-server at http://schloro.biocomp.unibo.it.
Funding
This work was partially supported by: PRIN 2010-2011 project 20108XYHJS (to P.L.M.) (Italian MIUR); COST BMBS Action TD1101 and Action BM1405 (European Union RTD Framework Program, to R.C); PON projects PON01_02249 and PAN Lab PONa3_00166 (Italian Miur to R.C.and P.L.M.); FARB UNIBO 2012 (to R.C.).
Conflict of Interest: none declared.
References
- Altschul S.F. et al. (1997) Gapped BLAST and PS I-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce B.D. (2001) The paradox of plastid transit peptides: conservation of function despite divergence in primary structure. Biochim. Biophys. Acta Mol. Cell Res., 1541, 2–21. [DOI] [PubMed] [Google Scholar]
- Chang C.C. et al. (2011) LIBSVM: a library for support vector machines. ACM Trans. Intell. Syst. Technol., 2, 1–27. [Google Scholar]
- Chou K.C. (2001) Prediction of protein cellular attributes using pseudo-amino acid composition. Proteins Struct. Funct. Genet., 43, 246–255. [DOI] [PubMed] [Google Scholar]
- Cooper G.M., Hausman R.E. (2009) The cell: a molecular approach, 5th edition, Sinauer Associates Inc., Sunderland, MA. [Google Scholar]
- Du P. et al. (2009) SubChlo: predicting protein subchloroplast locations with pseudo-amino acid composition and the evidence-theoretic K-nearest neighbor (ET-KNN) algorithm. J. Theor. Biol., 261, 330–335. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O. et al. (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc., 2, 953–971. [DOI] [PubMed] [Google Scholar]
- Goldberg T. et al. (2014) LocTree3 prediction of localization. Nucleic Acid Res., 42, W350–W355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Yan X. (2012) BS-KNN: An effective algorithm for predicting protein subchloroplast localization. Evol. Bioinf., 2011, 79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai K., Nakai K. (2010) Prediction of subcellular locations of proteins: Where to proceed? Proteomics, 1010, 3970–3983. [DOI] [PubMed] [Google Scholar]
- Kawashima S. et al. (2008) AAindex: amino acid index database, progress report 2008. Nucleic Acids Res., 36, D202–D205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R. et al. (2014) Protein sub-nuclear localization prediction using svm and pfam domain information. PLoS One, 9, e98345.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R.F. (1982) A simple method for displaying the hydropathic character of a protein. J. Mol. Biol., 157, 105–132. [DOI] [PubMed] [Google Scholar]
- Lin H. et al. (2013) Using over-represented tetrapeptides to predict protein submitochondria locations. Acta Biotheor., 61, 259–268. [DOI] [PubMed] [Google Scholar]
- Marcotte E.M. et al. (2000) Localizing proteins in the cell from their phylogenetic profile. Proc. Nat. Acad. Sci. U. S. A., 97, 12115–12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair R., Rost B. (2005) Mimicking cellular sorting improves prediction of subcellular localization. J. Mol. Biol., 348, 85–100. [DOI] [PubMed] [Google Scholar]
- Nakai R., Horton P. (1999) PSORT: a program for detecting the sorting signals of proteins and predicting their subcellular localization. Trends Biochem. Sci., 24, 34–35. [DOI] [PubMed] [Google Scholar]
- Patron N.J., Waller R.F. (2007) Transit peptide diversity and divergence: a global analysis of plastid targeting signals. BioEssays, 29, 1048–1058. [DOI] [PubMed] [Google Scholar]
- Savojardo C. et al. (2015) TPpred3 detects and discriminates mitochondrial and chloroplastic targeting peptides in eukaryotic proteins. Bioinformatics, 31, 3269–3275. [DOI] [PubMed] [Google Scholar]
- Schleiff E., Becker T. (2010) Common ground for protein translocation: access control for mitochondria and chloroplasts. Nat. Rev. Mol. Cell Biol., 12, 48–59. [DOI] [PubMed] [Google Scholar]
- Schleiff E., Klösgen R.B. (2001) Without a little help from "my" friends: direct insertion of proteins into chloroplast membranes? Biochim. Biophys. Acta Mol. Cell Res., 1541, 22–33. [DOI] [PubMed] [Google Scholar]
- Shi S.P. et al. (2011) Identify submitochondria and subchloroplast locations with pseudo amino acid composition: Approach from the strategy of discrete wavelet transform feature extraction. Biochim. Biophys. Acta Mol. Cell Res., 1813, 424–430. [DOI] [PubMed] [Google Scholar]
- Soll J. (2002) Protein import into chloroplasts. Curr. Opin. Plant Biol., 5, 529–535. [DOI] [PubMed] [Google Scholar]
- The UniProt Consortium (2014) UniProt: a hub for protein information. Nucleic Acids Res., 43, D204–D212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung C.W. et al. (2010) Prediction of protein subchloroplast locations using random forests. World Acad. Sci. Eng. Technol., 65, 903–907. [Google Scholar]
- Wang X. et al. (2015) MultiP-SChlo: multi-label protein subchloroplast localization prediction with Chou’s pseudo amino acid composition and a novel multi-label classifier. Bioinformatics, 31, 2639–2645. [DOI] [PubMed] [Google Scholar]