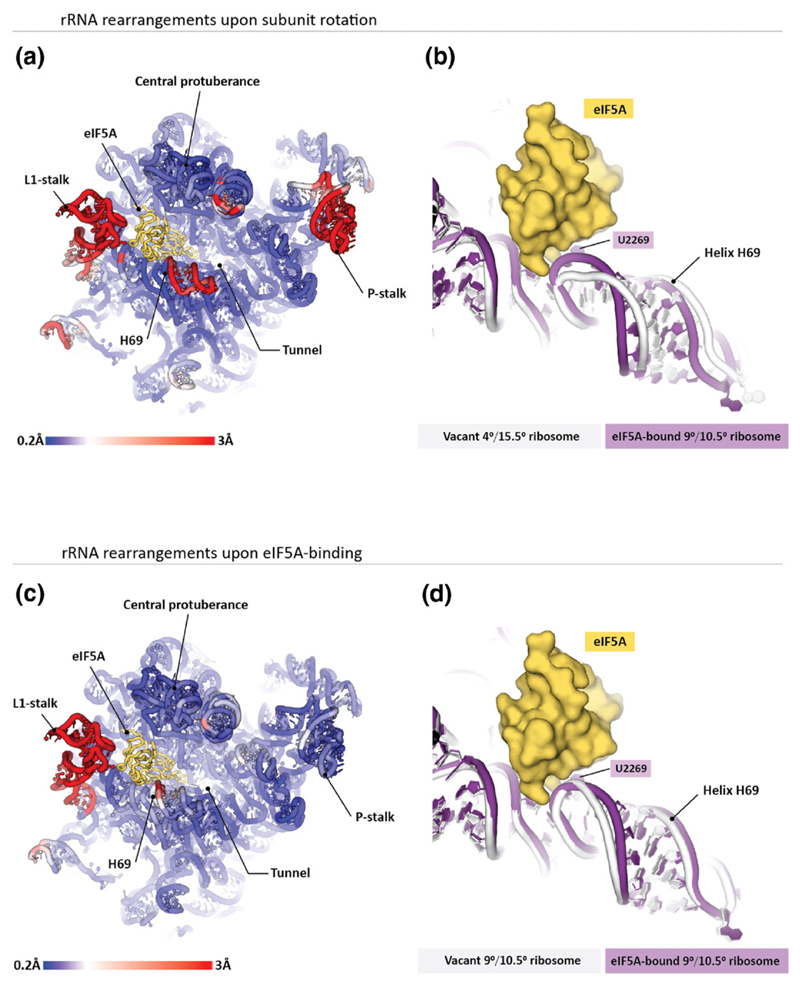
Fig. 3. Structure of the eIF5A-binding site depends on the ribosome rotation state. Panels (a and b) illustrate that ribosome transitions between the rotated states cause marked changes in the eIF5A-binding site.
(a) The 25S rRNA from the S. cerevisiae 80S ribosome is colored by displacement of the corresponding nucleotides between the 9°/10.5° (40S rotation/40S head swivel; eIF5A-bound) and 4°/15.5° (no eIF5A) rotated states of the two ribosomes in the asymmetric unit. The most prominent difference in the eIF5A-binding site is observed in helix H69. (b) Zoom on helix H69 shows that in the eIF5A-bound ribosome, helix H69 directly contacts eIF5A, whereas in the vacant ribosome, helix H69 moves ~6 Å away when eIF5A is docked in the equivalent position. Panels (c and d) illustrate structural changes in the 25S rRNA upon eIF5A binding to the ribosome. Two crystal structures of the yeast 80S ribosome are compared: the vacant 4°/15.5°-rotated ribosome (pdb 4v7r) and the eIF5A-bound 4°/15.5°-rotated ribosome (this study). (c), The 25S rRNA from the S. cerevisiae 80S ribosome is colored as in (a). (d) Zoom on helix H69 illustrates that eIF5A binding induces relatively subtle changes in the helix H69 structure.