Abstract
Invasive aspergillosis by Aspergillus fumigatus is a leading cause of infection-related mortality in immune-compromised patients. In order to discover potential genetic targets to control A. fumigatus infections we characterized rtfA, a gene encoding a putative RNA polymerase II transcription elongation factor-like protein. Our recent work has shown that the rtfA ortholog in the model fungus Aspergillus nidulans regulates morphogenesis and secondary metabolism. The present study on the opportunistic pathogen A. fumigatus rtfA gene revealed that this gene influences fungal growth and conidiation, as well as production of the secondary metabolites tryptoquivaline F, pseurotin A, fumiquinazoline C, festuclavine, and fumigaclavines A, B and C. Additionally, rtfA influences protease activity levels, the sensitivity to oxidative stress and adhesion capacity, all factors important in pathogenicity. Furthermore, rtfA was shown to be indispensable for normal virulence using Galleria mellonella as well as murine infection model systems.
Introduction
Aspergillus fumigatus has transitioned from being considered a saprophytic fungus of minor interest to recognition as one of the most important fungal pathogens, causing up to 90% of systemic Aspergillus infections [1]. This fungus produces spores, denominated conidia or conidiospores, which are ubiquitously present in the air and are capable of reaching the lung alveoli, where the infection is initiated [2, 3]. The spores of A. fumigatus range between 2–3 μm in diameter compared to the 3.5–8 μm diameter of other Aspergilli [2, 4]. The smaller size allows the conidia to evade capture by cilia and the mucosa [2]. Immunocompromised individuals are the main at-risk group for this fungal infection. This group includes patients who have received transplants, cancer patients undergoing chemotherapy and those with hematological malignancies or AIDS [5–10].
Aspergillus fumigatus infections can result in aspergillomas; mycelial balls established in pre-existing lung cavities. Patients with aspergillomas are usually asymptomatic and are only diagnosed when chest radiographs are taken for other medical complications [11]. In addition, there are several types of allergic pulmonary diseases caused by Aspergillus species. The most severe of these is Allergic Bronchopulmonary Aspergillosis (ABPA). This pulmonary disease occurs when A. fumigatus colonizes the bronchi in patients already affected by asthma or cystic fibrosis [12–14]. The complications incurred in this disease state range from exacerbation of asthma to fatal destruction of the lungs [15, 16].
However, the most severe condition caused by A. fumigatus infection is invasive aspergillosis (IA), with a mortality rate of 40–90% in immunocompromised patients [1, 11, 17–19]. Specific diagnostic methods as well as treatments are still limited, and in many cases not effective [20]. The prevalence and often-fatal effects of IA demands further investigation to gain insight into A. fumigatus infection traits that could provide the basis for the development of novel successful therapeutic approaches, for instance, A. fumigatus virulence factors including secondary metabolites, proteases, or conidial cell components [21].
The present study includes the characterization of the rtf1 putative ortholog in A. fumigatus. An rtf1 homolog has been described by Stolinski and collaborators in Saccharomyces cerevisiae [22]. It encodes a protein that functions as an RNA polymerase II transcription elongation factor. In that study, rtf1 was described to have a role in transcription initiation by regulating the binding of TATA box-binding protein (TBP) to the TATA element of the promoter [22]. Subsequent studies have assigned additional functions for rtf1 in yeast, including chromatin modifications. The Rtf1 protein has been shown to regulate the mono-ubiquitination of histone H2B at lysine 123 [23], as well as being key to recruit the chromatin remodeler Chd1 [24]. Further studies in both yeast and humans indicated that Rtf1 has chromatin-binding capabilities [24, 25]. Homologs of rtf1 have been found in many organisms. For instance, in the zebra fish, Danio rerio, mutations in this gene cause improper organ development of the neural crest. Also, in mice, rtf1 is required for proper embryonic stem cell identity [26, 27].
Previously, our laboratory characterized the rtf1 ortholog in the model filamentous fungus Aspergillus nidulans, where this gene regulates asexual and sexual development as well as secondary metabolism, including the production of the sterigmatocystin toxin [28]. In addition, our recent study of an rtfA homolog in Aspergillus flavus also revealed that a loss of rtfA results in a reduction in conidial production and sclerotial formation, accompanied by a reduction in the synthesis of the carcinogenic compound aflatoxin B1 [29]. rtfA is conserved in numerous fungal species, including A. fumigatus [28].
Our present study revealed that rtfA is necessary for proper growth as well as conidiation in A. fumigatus. In addition, secondary metabolite production was also influenced by rtfA, specifically production of festuclavine, fumigiclavines A, B, and C, pseurotin A, fumiquinazoline C, and tryptoquivaline F. Concomitantly, the expression of fgaPT2, psoA, and fmqA was also influenced by rtfA. Furthermore, rtfA was indispensable for full pathogenicity in Galleria mellonella and murine infection models. Other virulence factors, such as protease activity and resistance to oxidative stress were also reduced in the absence of rtfA in this opportunistic human pathogen.
Methods and materials
Generation of rtfA deletion strain
To construct the rtfA deletion strain (ΔrtfA), a deletion cassette was generated using fusion PCR as previously described by Szewczyk and collaborators [30]. The rtfA 5’ and 3’ UTRs were amplified from A. fumigatus genomic DNA using primers AfumRM3_5f and AfumRM3_5r and primers AfumRM3_3f and AfumRM3_3r, respectively (primers used in this study are included in S1 Table). A. parasiticus pyrG (2645 bp) selection marker was amplified from A. parasiticus genomic DNA with primers AparapyrG_fumRM3_f and AparapyrG_fumRM3_r. The 5’ and 3’ UTRs were fused to A. parasiticus pyrG using primers AfumRM35_nested and AfumRM33_nested. Aspergillus fumigatus CEA17 strain was transformed with the deletion cassette to replace the rtfA coding region with pyrG. Mutant colonies were grown on solid Glucose Minimal Medium (GMM) at 37°C. Genomic DNA from selected transformants was extracted for diagnosis. Deletion of rtfA was confirmed by Southern blot analysis. Quantitative reverse transcriptase PCR (qRT-PCR) was also used to confirm the absence of rtfA expression. The rtfA deletion strain was named TRRM4 (S2 Table).
Generation of a complementation strain
Creation of the rtfA complementation strain, TRRM5, began by generating a complementation plasmid, pRRM3. A 5750 bp fragment that included rtfA and the flanking UTRs was amplified from CEA10 using primers AfRM3_compFNot1 and AfRM3_compRAsc1 (S1 Table), which was then ligated into pJET cloning vector (ThermoFisher, Waltham, MA). Then the insert was released by HindIII digestions and ligated to pTDS3 [31], previously digested with the same enzyme. pTDS3 vector contains a pyrithiamine resistance selection marker. The resulting vector was denominated pRRM3. TRRM4 was transformed with pRRM3, resulting in the complementation strain TRRM5 (S2 Table). This strain was confirmed by diagnostic PCR using primers AfumRM3_Oef and AfumRM3_Oer.
Generation of an overexpression strain
A strain overexpressing rtfA was generated by transforming A. fumigatus CEA17 with the pRRM2 plasmid. To generate pRRM2, the rtfA coding sequence was first amplified from A. fumigatus CEA10 using primers AfumRM3_Oef and AfumRM3_Oer (S1 Table), which were engineered with AscI and NotI restriction sites, respectively. The rtfA PCR product was digested with AscI and NotI and ligated to pTDS1 [31], previously digested with the same enzymes. The resulting plasmid was named pRRM2. It contains the A. fumigatus pyrG selection marker and a gpdA promoter to induce constitutive expression. Transformants were screened by PCR using primers gdpApromoF and AfumRM3_Oer. The overexpression strain was named TRRM2 (S2 Table).
Generation of a heterologous complementation strain
The Saccharomyces cerevisiae RTF1 coding sequence was PCR amplified from genomic DNA from the yeast strain Y2HGold (Clontech, Mountain View, CA) using primers Scer_rtf1_Afum_rtfA_f and Scer_rtf1_Afum_rtfA_r (S1 Table) and fused to the 5’ and 3’ UTRs of A. fumigatus rtfA using fusion PCR as previously described [30]. The three fragments were fused using primers Afum_rtfA_nested_f and Afum_rtfA_nested_f, engineered with NotI restriction sites. This cassette was digested with NotI and ligated to pTDS3 pre-digested with the same enzyme. The resulting vector was denominated pRRM4. This plasmid was then transformed into A. fumigatus TRRM3. Confirmation of the colonies obtained by this heterologous complementation was accomplished by diagnostic PCR, using primers Scer_rtf1_Afum_rtfA_f and Scer_rtf1_Afum_rtfA_r, as well as by qRT-PCR with primers Scer_rtf1_qPCR_F and Scer_rtf1_qPCR_R. The heterologous strain was denominated TRRM7 (S2 Table).
Fluorescence microscopy
Aspergillus fumigatus ΔKU80 was transformed with an rtfA::gfp::pyrG fusion PCR cassette generated as previously described by Szewczyk [30]. Briefly, rtfA was amplified from CEA10 with primers AfumRM3_Oef and afumrm3r, excluding the stop codon. A gfp::pyrG fragment was PCR-amplified from plasmid p1439 using primers afumrm3gfpf and afumrm3gfpr (S1 Table). The rtfA 3’UTR was amplified from CEA10 with primers AfumRM3_3f and AfumRM3_3r. The three fragments were fused using primers afumrm3f and AfumRM33_Nested. The GFP-tagged transformant strain (TRRM6, S2 Table) was confirmed by PCR using primers AfumRM3_oef and AfumRM33_Nested. Conidia from TRRM6 were inoculated on the surface of coverslips immersed in Watch minimal medium [32]. The cultures were incubated for 16 h at 37°C. Localization of RtfA was visualized by observing green fluorescence (GF) using a Nikon Eclipe E600 microscope with a 60x immersion objective, Nomarski optics, and fluorochromes for green fluorescence detection (excitation, 470; emission, 525). GF images as well as differential interference contrast (DIC) and DAPI (4′,6-diamidino-2-phenylindole) images to indicate nuclear localization were obtained. Micrographs were captured with a Hamamatsu ORCA-ER digital camera processed by Hamamatsu HC Image software.
Gene expression analysis
Total RNA was extracted from lyophilized mycelium using Trizol (Invitrogen, Waltham, MA), following the manufacturer’s instructions. Gene expression was evaluated by qRT-PCR. For qRT-PCR, 2 μg of total RNA was treated with RQ1 RNase-Free DNase (Promega, Fitchburg, WI). cDNA was synthesized with Moloney murine leukemia virus (MMLV) reverse transcriptase (Promega, Madison, WI). qRT-PCR was performed with the Applied Biosystems 7000 Real-Time PCR System using SYBR green Jumpstart Taq Ready mix (Sigma, St. Louis, MO) for fluorescence detection. The primer pairs used for qRT-PCR are listed in S1 Table. The expression data for each gene was normalized to the A. fumigatus 18S RNA gene expression and the relative expression levels were calculated using the 2−ΔΔCT method [33].
Morphological analysis
For assessment of the effect of rtfA on colony growth, A. fumigatus wild type, ΔrtfA, complementation, and OErtfA strains were point-inoculated on solid GMM and incubated at 37°C. Colony diameter was measured after 5 days. The experiment was carried out with three replicates.
To assess the role of rtfA in conidiation, the same set of A. fumigatus strains were inoculated in GMM medium (1x106 spores ml-1) and incubated at 37°C as liquid stationary cultures. Fungal mycelial mats were collected at 48 h and 72 h. Samples were homogenized and spores were quantified with a hemacytometer using a Nikon Eclipse E400 microscope. Mycelial samples were also collected for RNA extraction at 48 h and 72 h. The experiment was carried out with three replicates. In addition, conidiation was also assessed on solid GMM. After 6 days, 8 mm diameter cores were collected from the cultures 0.5 cm from the center of the colony and homogenized in water. Conidia were counted with a hemacytometer using a Nikon Eclipse E400 microscope.
Conidiation was also examined in submerged conditions. Flasks containing 50 ml liquid GMM were inoculated with conidia (1x107spores ml-1) of A. fumigatus wild type, ΔrtfA, complementation, and OErtfA strains. Liquid shaking cultures were incubated at 37°C and 250 rpm. Fungal pellets were viewed under a Nikon Eclipse E600 microscope 72 h after inoculation, and micrographs were captured and processed by a Nikon DS-Fi1c camera and Nikon NIS-elements software. The experiment included two replicates.
Protease activity
A. fumigatus wild type, ΔrtfA, complementation, and OErtfA strains were point-inoculated on GMM medium (1% agar) and 5% skim milk (Difco, Sparks, MD) and incubated at 37°C in the dark. After four days, 72 mm diameter cores were collected from the cultures, blended in 30 ml distilled water and collected in 50 ml Falcon tubes. The tubes were centrifuged at 3,500 rpm at 4°C and 1 ml of supernatant was transferred to 1.5 ml microcentrifuge tubes and spun at 10,000 rpm for 10 min at 4°C. An Azo-Casein assay was performed as previously described by Reichard and collaborators [34] with slight modifications. One-hundred microliters of supernatant were mixed with 400 μl of Azocasein (Sigma, St. Louis, MO) at a concentration of 5 mg ml-1 dissolved in 50 mM Tris buffer (pH 7.5), 0.2 M NaCl, 5mM CaCl2, 0.05% Brij 35, and 0.01% sodium azide and incubated at 37°C for 90 min. One hundred and fifty microliters of 20% Trichloroacetic acid was then added to stop the reaction and the samples were left at room temperature for 30 min. The tubes were spun at 8,000 rpm for 3 min and 500 μl of the supernatant was mixed with 500 μl 1 M NaOH. Two hundred microliters from each sample was placed into a 96 well plate in duplicates and the absorbance of the released azo group was read at 436 nm using a plate reader (Epoch by Biotek). A negative control was used with sterile distilled water with azocasein.
Secondary metabolite extraction
Plates containing 25 ml liquid GMM were inoculated with 106 spores ml-1 of A. fumigatus wild type, ΔrtfA, complementation, and OErtfA strains and incubated at 37°C. Supernatant was collected at 120 h (three replicates) for secondary metabolite extraction and filtered through Miracloth (Calbiochem, Billerica, MA) into 50 ml conical centrifuge tubes. Twelve milliliters were collected from each replicate. Samples were extracted with an equal amount of chloroform. The bottom layer was then transferred to a glass beaker and allowed to dry. Extracts were resuspended in 1 ml methanol and then filtered through 0.22 μm diameter pore filters into 1.5 ml microcentrifuge tubes, where they were allowed to evaporate completely.
Liquid chromatography and mass spectrometry analysis
Sample analysis was performed using HPLC coupled to an LTQ Orbitrap XL high-resolution mass spectrometer (Thermo Fisher Scientific, Les Ulis, France). Extracts were resuspended in 400 μl methanol and 10 μL of this suspension were injected into a reversed-phase (150 mm × 2.0 mm) 5 μm Luna C18 column (Phenomenex, Torrance, CA, U.S.A.) operated at a flow rate of 0.2 mL/min. A gradient program was performed with 0.1% formic acid (phase A) and 100% acetonitrile (phase B) with the following elution gradient: 0 min 20% B, 30 min 50% B, from 35 to 45 min 90% B, from 50 to 60 20% B. HRMS acquisitions were achieved with electrospray ionization (ESI) in the positive and negative modes as follows: spray voltage +4.5 kV, capillary temperature 350°C, sheath gas (N2) flow rate 40 au (arbitrary units), auxiliary gas (N2) flow rate 6 au in the positive mode, and spray voltage −3.7 kV, capillary temperature 350°C, sheath gas (N2) flow rate 30 au, auxiliary gas (N2) flow rate 10 au in the negative mode. Full MS spectra were acquired at a resolution of 60,000 with a range of mass-to-charge ratio (m/z) set to 50−800. The identity of fungal products was confirmed by comparison either with HPLC-MS2 analysis of a standard compound or on the base of results obtained in Gauthier et al. [35] and Cano et al. [36].
Cell wall tests
To assess possible alterations of the cell wall integrity due to changes in the rtfA locus, the wild type, ΔrtfA, complementation and overexpression strains were point-inoculated on 25 ml 1% agar GMM supplemented with Congo Red at concentrations of 0, 30, 40, 50, and 60 μg ml-1 and incubated at 37°C. Photographs were taken at 48 h and 72 h.
In a similar experiment the same strain set was point-inoculated on 25 ml 1% agar GMM supplemented with SDS at concentrations of 0, 0.005, 0.01, and 0.02% SDS and incubated at 37°C. Colony diameter measurement were taken at 48 h and 72 h.
In addition, the strains were also tested in the presence of the anti-fungal nikkomycin Z. Strains were point-inoculated in a 24-well plate on 1% agar GMM supplemented with 0, 32, or 64 μg ml-1 Nikkomycin Z. Plates were incubated at 37°C and photographs were taken at 48 h after inoculation.
6-azauracil sensitivity
The wild type, ΔrtfA, complementation, and overexpression strains were point-inoculated on solid (1% agar) GMM plates supplemented with 6-azauracil at concentrations of 0, 50, 100, and 300 μg ml-1. Plates were incubated at 37°C and colony diameters were measured at 48 h and 72 h.
Adhesion capacity test
In order to evaluate the possible role of rtfA in adhesion capacity on inanimate surfaces, each strain was inoculated in 12 ml GMM at 105 spores/ml, homogenized and 130 μl of these suspensions was added to each well of a 96-well plate and incubated at 37°C for 24 h, 48 h, and 72 h. After incubation the medium was removed and the mycelia were washed three times with water. Then they were stained with 130 μl 0.01% Crystal Violet in water. Staining was allowed for 20 min at room temperature, washed three times with water, allowed to dry, and then destained with 130 μl of 30% acetic acid. The absorbance was read at 560 nm on an Epoch spectrophotometer (Biotek, Winooski, VT).
Environmental stress tests
All strains were point-inoculated on solid GMM (1% agar) and incubated at 37°C. Fungal growth was assessed as colony diameter and documented with photographs. Temperature stress tests were performed at 25°C, 30°C, 37°C, 42°C, and 45°C. Response to osmotic stress was analyzed by supplementing the medium with either 1.2 M sorbitol, 0.6 M KCl, or 1.0 M sucrose. Analysis of the rtfA role in response to pH stress was performed by adjusting the pH of the medium to 5, 6, 7 or 8.
To evaluate the importance of rtfA in the resistance to oxidative stress, the strains were point-inoculated on solid GMM plates supplemented with 0, 5, 10, 15, 20, 25, 30, or 35 μM menadione in a 24-well plate and incubated at 37°C for 48 h or 72 h. A separate experiment was done inoculating the strains (1 x 107 spores ml-1) in liquid GMM and growing at 30°C for 24 h. One gram of mycelium was then transferred to 50 ml liquid GMM with or without 20 μM menadione and incubated at 30°C and 200 rpm in a shaking incubator. Mycelium was collected after 6 h for gene expression analysis of cat1 and cat2.
Pathogenicity analysis in the Galleria mellonella model
Spore suspensions of A. fumigatus wild type, ΔrtfA, complementation and OErtfA strains were generated in 1x PBS (with 0.1% Tween). The spore suspensions were quantified using a hemacytometer and diluted in 1x PBS to a concentration of either 1x 105 or 1x106 spores 10 μl-1. The infection procedure was done as previously described by Fuchs [37]. Briefly, Galleria mellonella larvae (Vanderhorst Wholesale, Saint Marys, Ohio) with a weight range between 275–300 mg and lacking grey markings were selected for the experiment. Groups of 30 larvae were selected for each A. fumigatus strain. PBS injections and larvae without injections were used as controls. The larvae were injected with spores using a syringe behind their last left pro-leg. The larvae were transferred into a Petri dish (90 mm x 15 mm) and wrapped in aluminum foil. The plates were placed at 37°C in the dark. After 16 h, the larvae were checked every two hours for mortality. Mortality curves were generated using SPSS software and a log rank test was performed to generate pair-wise comparisons of the survival of the larvae infected with different strains. No ethical approval was required for this species because they are unregulated animals.
Pathogenicity analysis in a murine model
To examine the role of rtfA in an in vivo mammalian model, female 6-week-old, outbred Swiss ICR mice (Harlan Sprague Dawley, Indianapolis, IN), weighing 24 to 27 g, were immunosuppressed (neutropenic) by subcutaneous injection of cyclophosphamide (150 mg kg-1 of body weight) on days −4, −1, and 3 with a single dose of cortisone acetate (200 mg kg-1), given subcutaneously on the day of spore inoculation. The mice were anesthetized with isoflurane inhalation on day 0. The mice were infected by intranasal inoculation, mimicking the natural route of infection. Inoculum was prepared from three strains: wild type, ΔrtfA, and complementation strains grown on glucose minimal medium (GMM) plates. Conidia were harvested by flooding fungal colonies with 0.85% NaCl with Tween 80, enumerated with a hemocytometer, and adjusted to a final concentration of 1x106 spores ml-1. Sedated mice (10 mice per fungal strain) were infected by nasal instillation of 50 μl of the inoculum (day 1) and monitored three times daily for 7 days post-infection. All surviving mice were sacrificed on day 7. Cumulative mortality curves were generated, and the statistical analysis using the log rank test was utilized to perform pairwise comparisons of survival levels among the strain groups.
This study was carried out in strict accordance with the Guide for the Care and Use of Laboratory Animals of the National Research Council. The protocol was approved by the Institutional Animal Care and Use Committee of Northern Illinois University (Permit #12–0006). All efforts were made to minimize suffering. Humane euthanasia by C02 inhalation was performed when mice met criteria indicating a moribund state; these endpoints include behaviors of unresponsiveness to tactile stimuli, inactivity, lethargy, staggering, anorexia and/or clinical signs of bleeding from the nose or mouth, labored breathing, agonal respirations, purulent exudate from eyes or nose, abnormally ruffled fur, or greater than 20% weight loss. The method of euthanasia by CO2 inhalation is consistent with recommendations of the Panel on Euthanasia of the American Veterinary Medical Association.
Statistical analysis
All statistical analysis was completed using IBM SPSS software. Unless otherwise indicated, three replicates were used and a p value of 0.05 was used for statistical analysis. Error bars represent standard error.
Results
Identification of rtfA in A. fumigatus
Rtf1 presents the well-characterized conserved domain Plus3, which has homology to other nucleic acid binding proteins, including the PAZ domains on the endoribonucleases Dicer and Argonaute from Drosophila melanogaster as well as the KOW domain of the bacterial transcription elongation factor NusG; the Plus3 domain was demonstrated to bind to RNA polymerase II as well as to single-stranded DNA in humans [25]. The Plus3 domain receives its name from three conserved positively charged amino acids—two arginine residues and a serine [25]. The domain encompasses amino acids 244–343 in S. cerevisiae. A BlastP search (NCBI) using S. cerevisiae Rtf1 as query to find homologs in A. fumigatus revealed only one hit. The rtfA gene (Afu2g01900) consists of 2036 nucleotides located on the positive strand of chromosome 2. The A. fumigatus RtfA deduced encoded protein (accession number, XP 749325.1) is 607 amino acids long. A comparison between S. cerevisiae Rtf1 and A. fumigatus RtfA homologs showed 27% identity. Aspergillus nidulans RtfA also contains the conserved Plus3 domain, identified as amino acids 271–375. Comparison between RtfA from A. nidulans (AN4570) and its homolog in A. fumigatus revealed a 66% identity.
Growth rate and conidiation are regulated by rtfA
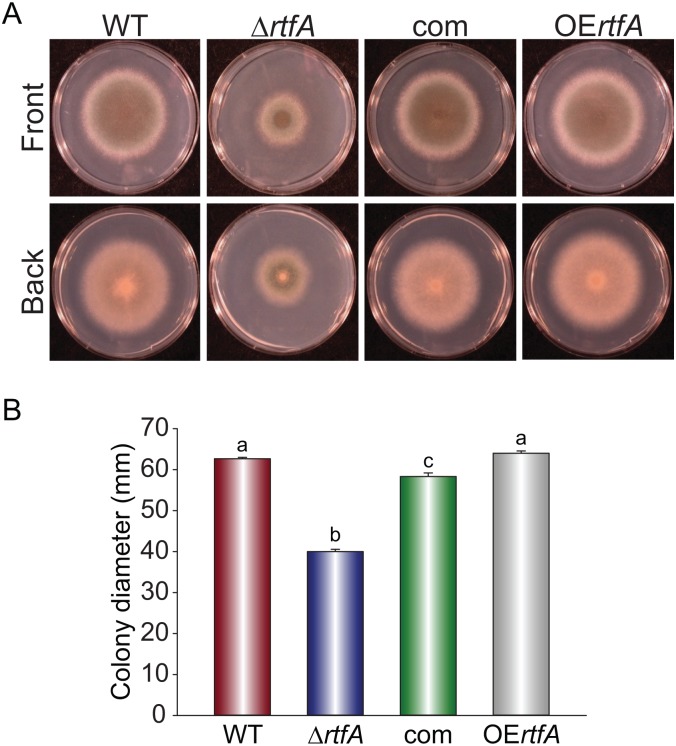
To determine the effects of rtfA on growth, conidiation, and other cellular processes, rtfA deletion (ΔrtfA), complementation, and overexpression strains were generated. The deletion strain was confirmed by Southern blot analysis (S1A Fig). Genomic DNA from both wild type and ΔrtfA strain was isolated and digested with Sal I. A 1624 kb DNA fragment corresponding to the 5’ UTR region of rtfA was used to generate the radioactive probe utilized in this hybridization. The presence of 5.1 and 3.7 kb bands in the Southern blot analysis confirmed the rftA deletion, while 5.1 and 1.6 kb bands corresponded to the wild type control (S1A Fig). With respect to the complementation strain, diagnostic PCR was used to verify the integration of the wild type allele in the ΔrtfA strain (S1B Fig). PCR was also utilized to verify the overexpression strains (S1C Fig). In addition, qRT-PCR was used to confirm the lack of rtfA expression in the ΔrtfA mutant under conditions that allow its transcription in the wild type and complementation control strain (S1D Fig). Forced high levels of rtfA transcripts in the overexpression strain (40x compared to wild type) were also confirmed by this method (S1D Fig). Examination of the colony growth rate of wild type and ΔrtfA strain revealed a significant decrease (37%) in colony diameter in the absence of rtfA compared to the wild type (Fig 1A and 1B). A green pigment appeared in the deletion mutant colony when viewed from beneath. This green pigment was absent in the isogenic wild type strain. Complementation of ΔrtfA with the rtfA wild type allele recovered wild type phenotype. Overexpression of rtfA did not show any effects on growth.
Fig 1. rtfA affects A. fumigatus colony growth.
Aspergillus fumigatus wild type (WT), ΔrtfA, complementation (com), and overexpression rtfA (OErtfA) strains were point-inoculated on GMM and incubated for 4 days at 37°C. (A) Photographs of the colonies. (B) Colony growth estimated as colony diameter. Different letters above the bars indicate significantly different values (p ≤ 0.05). Error bars represent standard error.
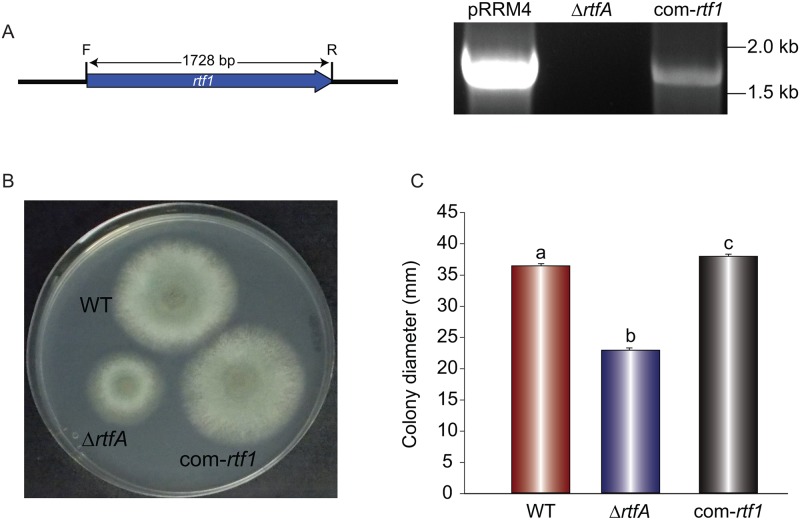
To examine possible functional similarity of A. fumigatus rtfA with that of S. cerevisiae rtf1, A. fumigatus ΔrtfA was complemented with rtf1 from yeast (Fig 2). After successful complementation, com-rtf1 (TRRM7) was compared to the wild type strain and the ΔrtfA strain. The three strains were incubated for 3 days on GMM. Our results showed that complementation with the S. cerevisiae genes rescued wild-type phenotype in A. fumigatus.
Fig 2. Heterologous complementation with S. cerevisiae rtf1 restores wild-type phenotype in the A. fumigatus rtfA mutant.
Aspergillus fumigatus ΔrtfA strain (TRRM3) was transformed with plasmid pRRM4, which contains rtf1 from S. cerevisiae. (A) Effective complementation with rtf1 was confirmed by diagnostic PCR analysis using primers Scer_rtf1Afum_rtfA_f and Scer_rtf1_Afum_rtfA_r. Plasmid pRRM4 was used as a positive control, yielding the expected 1.7 kb PCR product. Aspergillus fumigatus ΔrtfA strain was used as negative control. (B) A. fumigatus strains WT (CEA10), ΔrtfA (TRRM4), and com-rtf1 (TRRM7) were point-inoculated on GMM and incubated at 37°C. (C) Colony diameters were measured after three days of growth. Values represent the average of three replicates. Different letters above the bars indicate significantly different values (p ≤ 0.05). Error bars represent standard error.
Previous studies indicated that rtf1 is a transcription elongation factor [23, 24]. Cchemical treatment with 6-azauracil (6-AU) results in a reduction of intracellular GTP and UTP levels, which alone is not lethal, but in combination with mutations that affect transcriptional elongation, can prevent growth [38, 39]. Loss of rtf1 in S. cerevisiae significantly affected the ability of the yeast strain to grow in the presence of 6-AU [40]. We tested the A. fumigatus ΔrtfA strain’s ability to grow in the presence of this chemical. Increasing concentrations of 6-AU in the medium lead to a gradual decrease in growth in all the strains assayed. Addition of this compound to the ΔrtfA cultures only slightly aggravated the observed colony growth reduction in A. fumigatus (S2 Fig).
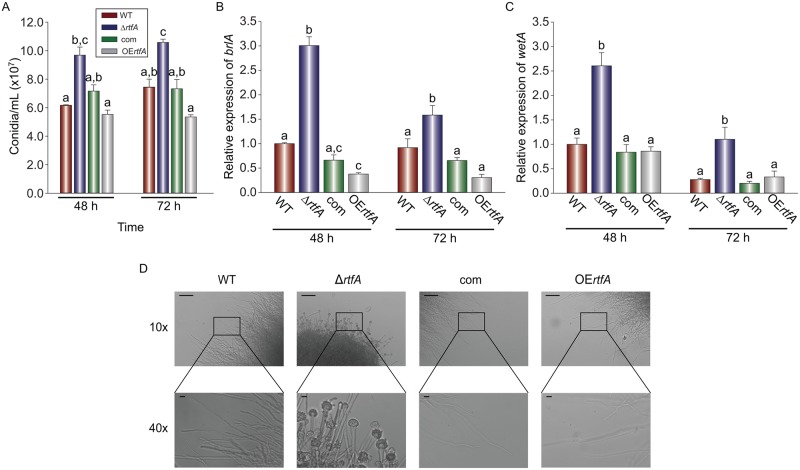
Airborne conidia are infectious inoculum for aspergillosis. Our previous studies indicated that production of these asexual spores was affected by rtfA in A. nidulans and A. flavus [28, 29]. In our current study, the role of rtfA in conidiation was also examined in A. fumigatus. The ΔrtfA strain showed a significant increase in conidial production compared to the wild type in liquid stationary cultures (Fig 3A). This hyperconidiation coincided with increased expression of two key regulatory genes in this developmental signaling pathway, brlA and wetA (Fig 3B and 3C) [41, 42]. Similarly, the rtfA deletion mutant also presented hyperconidiation when grown on solid medium (S3A Fig). Furthermore, conidiation was also observed in ΔrtfA in submerged cultures, a condition that inhibits conidiophore formation in the wild type (Fig 3D). Additionally, the ΔrtfA submerged culture accumulated a purple compound that was absent in the wild-type culture (S3B Fig).
Fig 3. rtfA is a repressor of conidiation in A. fumigatus.
(A) Wild type (WT), ΔrtfA, complementation (com), and overexpression rtfA (OErtfA) strains were grown in liquid stationary cultures and whole mycelial mats were collected at 48 h and 72 h, homogenized, and conidia were quantified with a hemocytometer. Expression analysis of brlA (B) and wetA (C), two key genes in the conidiation regulatory pathway, at 48 h and 72 h. (D) Micrographs showing conidiation in the ΔrtfA strain when grown in liquid shaking cultures. Cultures were grown at 37°C for 72 h at 250 rpm. Scale bars represent 100 μm in the 10x micrographs and 10 μm in the 40x micrographs.
RtfA subcellular localization
Previous work in A. nidulans revealed the nuclear localization of the RtfA homolog in this model fungus [28]. To investigate whether this holds true in A. fumigatus, we generated a strain with RtfA fused to GFP. The generated strain presented wild-type phenotype. Our results indicated that A. fumigatus RtfA is also localized in nuclei, as indicated by comparison with images corresponding to DAPI staining (S4 Fig).
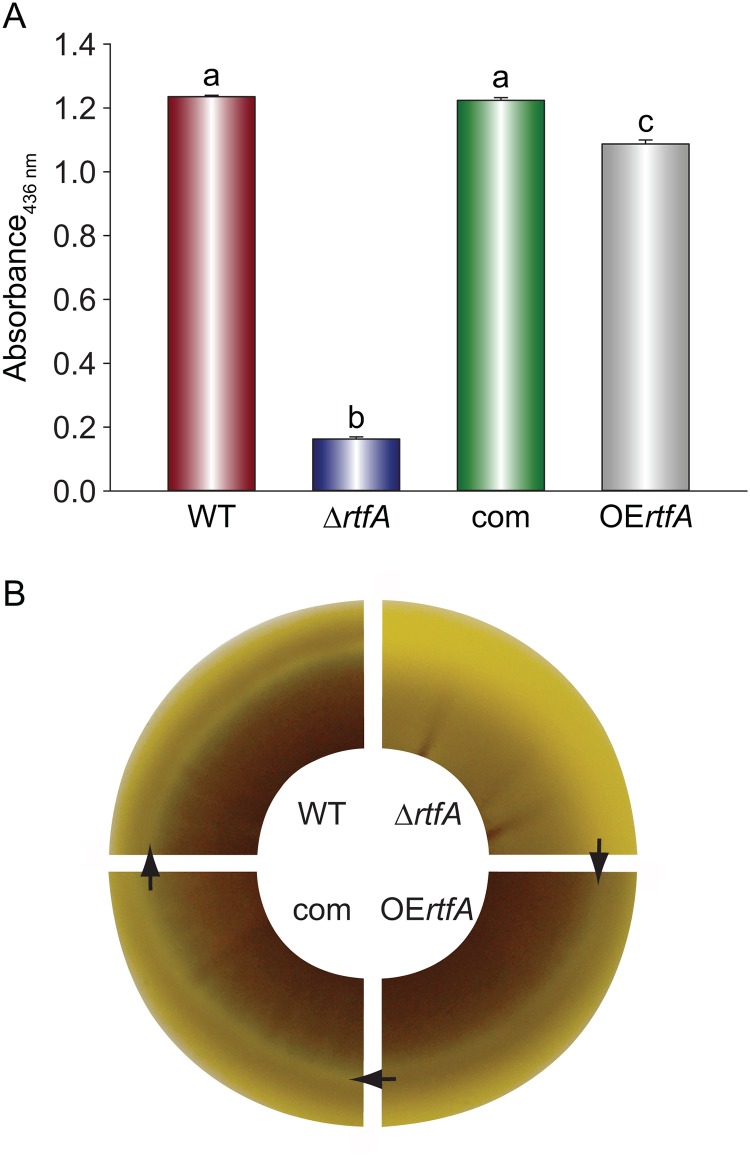
rtfA is required for normal protease activity
Studies in A. nidulans have shown that rtfA is functionally dependent on the global regulatory gene veA [28]. Our previous studies on A. fumigatus veA revealed that absence of veA or overexpression of this gene results in a decrease in protease activity levels [21]. It is possible that rtfA in A. fumigatus could also influence protease activity in this fungus, which has been associated with pathogenicity in some studies [43, 44] while not in others [45–47]. Similarly to the veA study, our current study revealed that alteration of the A. fumigatus rtfA locus results in variations in protease activity compared to those in the wild type (Fig 4A). Specifically, deletion of the rtfA gene led to a significant decrease of this hydrolytic activity when the cultures were grown on 5% skim milk agar plates. The proteolytic activity in cultures can be also visualized by degradation halos forming at the edge of the colonies. The degradation halo was absent in the deletion mutant, while they were visible in the other strains assayed (Fig 4B).
Fig 4. rtfA regulates protease activity.
Aspergillus fumigatus wild type (WT), ΔrtfA, complementation (com), and overexpression rtfA (OErtfA) strains were point-inoculated on GMM containing 5% skim milk (Difco) and incubated at 37°C. (A) Quantification of proteolytic activity is shown as measured by Azo-casein assay after four days of growth. Different letters above the bars indicate significantly different values (p ≤ 0.05). Error bars represent standard error. (B) Degradation halos (indicated with arrows) at the edge of the colonies growing on skim milk agar plates for four days.
rtfA influences A. fumigatus oxidative stress sensitivity
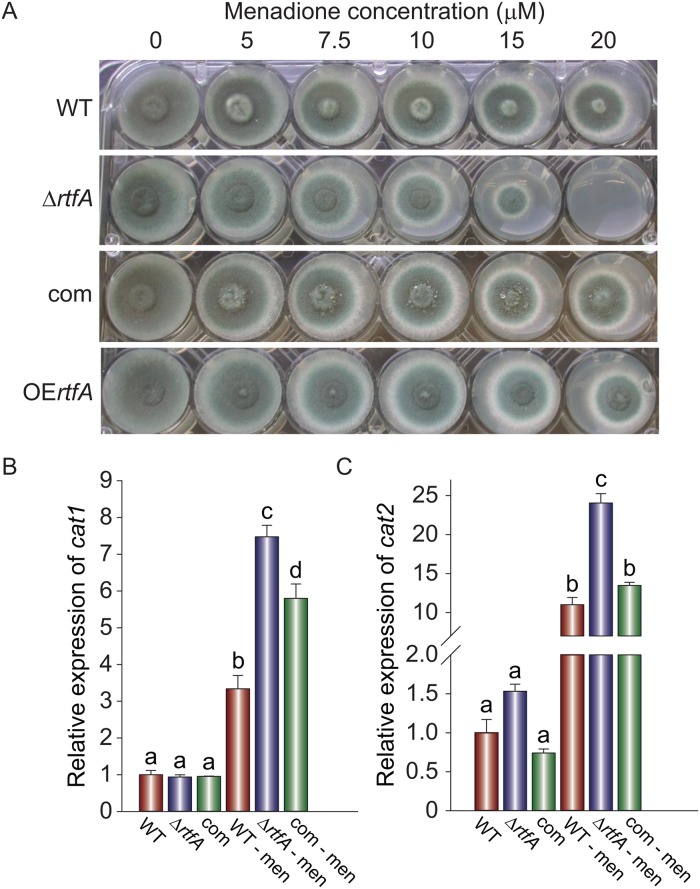
Generation of an oxidative stress environment is an important line of host defense against A. fumigatus infections [48, 49]. Our study showed that deletion of rtfA results in greater sensitivity to oxidative stress, being unable to grow in the presence of 20 μM menadione, a condition that allowed near-normal growth of wild type colonies (Fig 5). Overexpression of rtfA did not change the sensitivity to menadione compared to that in the wild type (Fig 5A and S5 Fig). In a separate experiment, the strains were also grown in liquid shaking cultures with or without menadione. Under these conditions, the expression of two genes involved in oxidative stress response, cat1 and cat2 [49] was unexpectedly higher in the ΔrtfA strain compared to the wild type after 6 h of exposure to menadione (Fig 5B and 5C). Absence or overexpression of rtfA did not alter the effect of other stress factors such as pH, temperature or osmostress on the growth of A. fumigatus (S6, S7, and S8 Figs).
Fig 5. rtfA is necessary for normal resistance to oxidative stress.
(A) Sensitivity of A. fumigatus wild type (WT), ΔrtfA, complementation (com), and overexpression rtfA (OErtfA) strains to oxidative stress was examined on GMM containing various concentrations of menadione as indicated. Colony formation was observed after 72 h of incubation at 37°C. (B, C) In a separate experiment, the strains were inoculated in liquid GMM with or without 20 μM of menadione (men). Cultures were incubated at 30°C. Mycelia were collected 6 h after the shift for expression analysis of cat1 (B) and cat2 (C). Different letters above the bars indicate significantly different values (p ≤ 0.05). Error bars represent standard error.
rtfA regulates A. fumigatus secondary metabolism
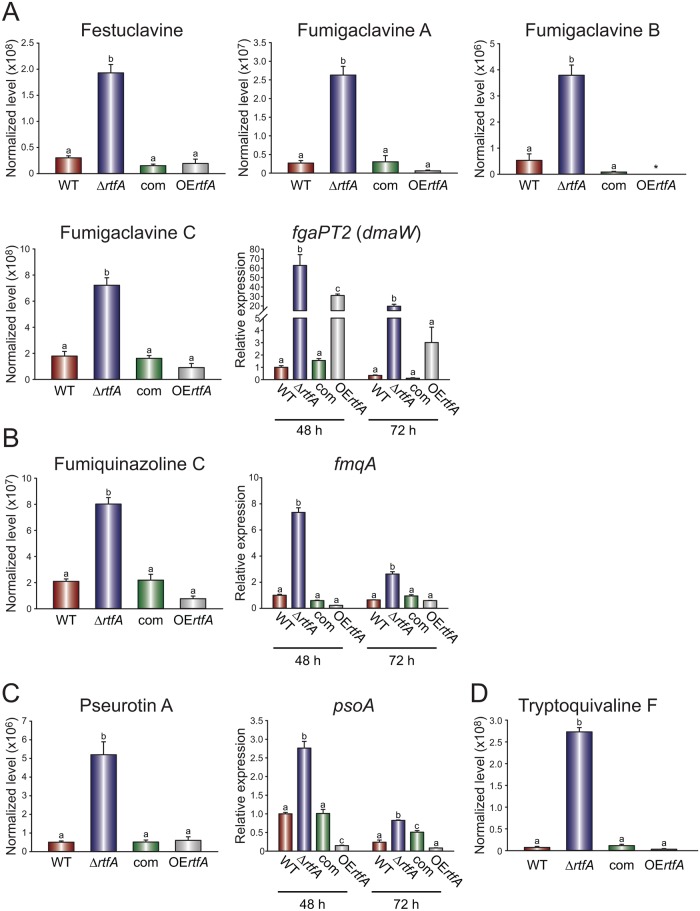
Aspergillus fumigatus produces a myriad of secondary metabolites with bioactive properties that serve multiple functions [50], including pathogenicity. We examined whether an assortment of secondary metabolites are rtfA-dependent in this fungus. Our LC-MS analysis revealed that various ergot alkaloids, including festuclavine and fumigaclavines A, B, and C, belonging to the same biosynthetic pathway, were produced at significantly higher levels in the rtfA mutant strain compared to the wild type (Fig 6A). Overexpression of rtfA resulted in undetectable production of fumigaclavine B (Fig 6A). Concomitantly, the gene fgaPT2, encoding an enzyme involved in an early step of this pathway [51], was expressed at higher levels in the ΔrtfA strain with respect to the controls at both 48 h and 72 h (Fig 6A). Interestingly, expression of fgaPT2 was also elevated in the overexpression strain compared to wild-type levels, however this increase was not sufficient to increase production of these alkaloids (Fig 6A).
Fig 6. rtfA regulates the production of secondary metabolites.
Plates containing 25 mL liquid GMM were inoculated with 106 spores/mL of A. fumigatus wild type (WT), deletion (ΔrtfA), complementation (com), and overexpression (OErtfA) strains. Cultures were incubated at 37°C. Mycelia were collected at 48 h and 72 h for expression analysis. Supernatant was collected at 120 h for secondary metabolite extraction and analysis. (A) Analysis of ergot alkaloids and fgaPT2 (dmaW), a key gene in this cluster. (B) Analysis of fumiquinazoline C and fmqA, the gene encoding a NRPS involved in fumiquinazoline C biosynthesis. (C) Analysis of pseurotin A and psoA, encoding the PKS-NRPS in this biosynthetic pathway. (D) Analysis of tryptoquivaline F production. Different letters above the bars indicate significantly different values (p ≤ 0.05). Error bars represent standard error.
The synthesis of another secondary metabolite, fumiquinazoline C, was also rtfA-dependent; production of this compound increased in the absence of rtfA (Fig 6B). Expression analysis of the fmqA gene, encoding the non-ribosomal peptide synthase (NRPS) in the fumiquinazoline C biosynthetic pathway [52], revealed an increase in transcription in the rtfA mutant, while a decrease was observed in the overexpression strains (Fig 6B). Similar results were observed when analyzing levels of pseurotin A production, as well as the expression of the psoA gene (Fig 6C), encoding a polyketide synthase-NRPS (PKS-NRPS) involved in the pseurotin A biosynthesis [53]. Furthermore, deletion of rtfA also drastically increased the production of tryptoquivaline F (Fig 6D).
rtfA effect on cell wall and adhesion capacity
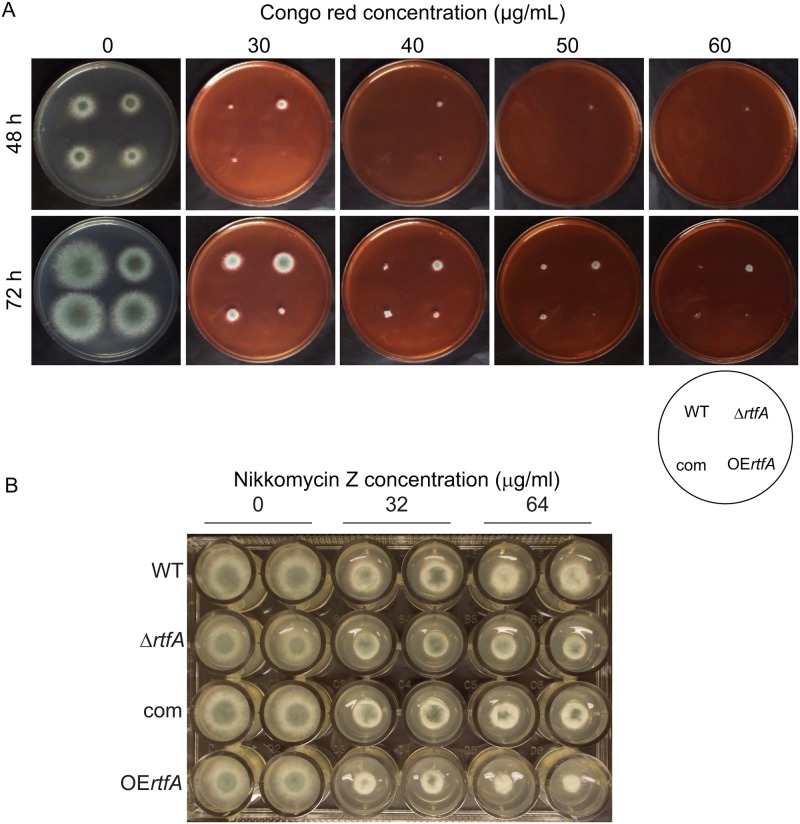
Aspergillus fumigatus cell wall represents the first point of contact with the host. To investigate the possible role of A. fumigatus rtfA in cell wall integrity, we subjected ΔrtfA and rtfA overexpression strains as well as their controls, wild type and complementation strains, to various known cell wall stressors commonly used in S. cerevisiae and Aspergillus studies [54–59]. When grown on medium supplemented with SDS, no noticeable differences were observed (S9 Fig). However, when Congo red or the chitin synthase inhibitor nikkomycin Z [56] were added to the medium, the overexpression strain presented an increase in sensitivity to these agents with respect to the control, while the rtfA mutant showed a slight increase in resistance, particularly in the presence of Congo red (Fig 7A and 7B).
Fig 7. Alteration of rtfA expression has a minor effect on cell wall integrity.
Wild type (WT), deletion (ΔrtfA), complementation (com), and overexpression (OErtfA) strains were point-inoculated on GMM supplemented with increasing concentrations of (A) Congo Red or (B) Nikkomycin Z as indicated. Strains were incubated at 37°C for 72 h (A) or 48 h (B).
In the host biofilm formation has been shown to be important in A. fumigatus resistance to anti-fungal drugs [60, 61, 62]. Adhesion to surfaces is necessary for the formation of biofilm [63, 63]. Our analysis shows an initial delay in the capacity of this fungus to bind to abiotic surfaces in the absence of rtfA. A reduction in adhesion was observed in this mutant at 24 h and 48 h after inoculation (S10 Fig). However, at 72 h, the ΔrtfA strain showed wild-type levels of this capacity.
Deletion of rtfA leads to a decrease in virulence in both Galleria mellonella and murine infection models
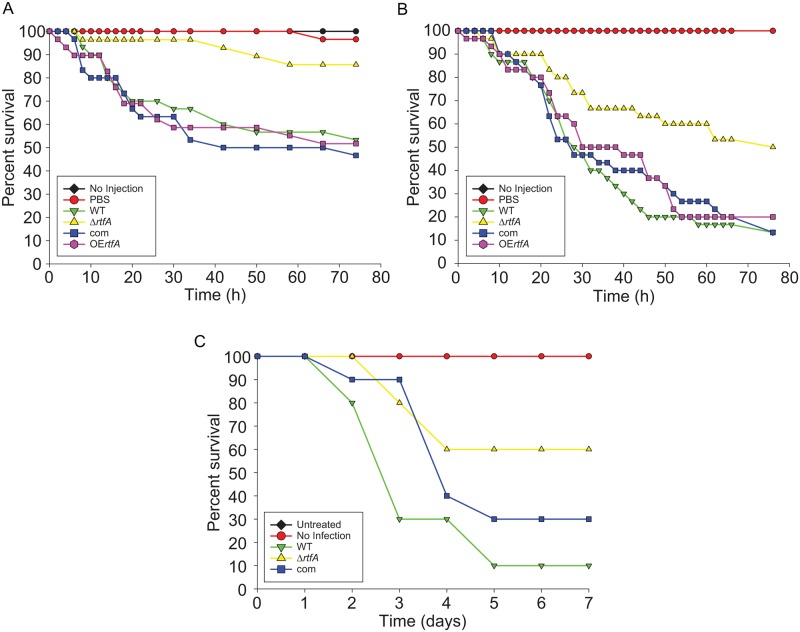
Since rtfA affects growth, conidiation, protease activity, production of secondary metabolites, and response to cell wall and oxidative stresses, it is likely that rtfA may play an important role in pathogenicity. To test this possibility Galleria mellonella larvae were infected with 105 spores/larva of wild type, ΔrtfA, complementation, or OErtfA strains. After 74 hours of observations, only 51% of the larvae infected with wild type survived, while 85% of the larvae infected with ΔrtfA were still alive (Fig 8A). Larvae infected with complementation and OErtfA strains showed similar survival rates to that of the wild type. In a similar experiment the larvae were also inoculated with an increased inoculum (106 spores/larva). Under these conditions 87% of the larvae infected with the wild type spores died, and those infected with the complementation and overexpression strains had a similar result, while 50% of the larva infected with the deletion mutant died (Fig 8B). The differences between the wild type and deletion mutant were statistically significant for both inoculum concentrations tested.
Fig 8. rtfA is indispensable for normal pathogenicity.
Larvae of G. mellonella were infected with conidia of A. fumigatus wild type (WT), ΔrtfA, complementation (com), and overexpression rtfA (OErtfA) strains as described in the experimental procedures section. Thirty larvae were infected for each group, including a control group injected with 1X PBS. Survival was first monitored every two hours and eventually extended to every 4 or 8 hours when the larval mortality rate decreased. (A) Larvae infected with 105 spores and (B) 106 spores. A no injection control and a PBS control were included. (C) Six-week old mice were rendered neutropenic by treatment with a cyclophosphamide and kenalog-10. Neutropenic mice were infected with 106 conidia per mouse of wild type (WT), ΔrtfA, or complementation strains and monitored daily for a total of seven days. Two controls were included, one with a group of non-infected mice rendered neutropenic and a second group with mice neither infected with spores nor treated with immunosuppressants. Statistical analysis was carried out by pairwise comparison using a long rank test.
Considering the results with the G. mellonella model, pathogenicity tests were then performed in a mammalian model (Fig 8). A sample size of 10 mice was used per strain. Similar to the results of the pathogenicity tests using the Galleria infection model, the mice infected with the mutant strain showed an increased survival rate compared to those infected with the wild-type strain; at the seventh day in this experiment, one mouse of the group infected with the A. fumigatus wild-type strain was still viable, while six mice were still alive in the group of animals infected with the rtfA mutant. Two control groups were used in this experiment. One control group was immunosuppressed but not infected with spores, while the other control group was neither immunosuppressed nor infected with spores. Both groups showed 100% survival.
Discussion
Aspergillus fumigatus rtfA encodes a nuclear putative RNA polymerase II transcription elongation factor-like protein that is homologous to rtf1 in Saccharomyces cerevisiae. Rtf1 is a member of a protein complex called Paf1C that includes the proteins Paf1, Cdc73, Ctr9, and Leo1. Originally thought to simply be an alternative for the Mediator protein complex, a coregulator of transcription [64], subsequent research has shown that the Paf1C associates with the RNA polymerase II from the pre-initiation complex to the 3’ end, where RNA is cleaved from the DNA template [22, 65, 66]. Indeed, Rtf1 has been shown to be the primary agent in binding the Paf1C to RNA Polymerase II [25]. The most studied role of the Paf1C is in the regulation of chromatin structure and histone modifications. In S. cerevisiae, Rtf1 is involved in regulating the mono-ubiquitylation of histone H2B at lysine 123, which is required for the methylation of histones H3K4 and H3K79 [23, 24, 67–71]. Interestingly, complementation of the A. fumigatus rtfA mutant with the S. cerevisiae rtf1 gene rescued wild-type phenotype, suggesting that both homologs carry out conserved functions. The fact that the 6-AU treatment did not affect the ΔrtfA strain in a similar manner as loss of rtf1 in S. cerevisiae [38, 40] does not necessarily indicate a different function for rtfA, since mutations in transcription elongation factors do not always bestow sensitivity to 6-AU [72]. It is possible that the RNA Polymerase II could rely more heavily on factors that promote elongation in yeast [24]. This A. fumigatus study, as well as previous studies in other Aspergillus [28, 29] and in yeast [i.e. 22, 65–71], revealed a broad regulatory role for rtfA, where this transcriptional regulator controls the expression of many genes, including those involved in conidiation. As previously mentioned, the main route of entry for A. fumigatus into humans is through inhalation of conidia [11]. Indeed, the conidia produced by A. fumigatus are ubiquitous in the air and it is estimated that humans inhale a few hundred conidia per day [48]. Our results indicate that rtfA is involved in the regulation of conidial production. Deletion of rtfA resulted in hyperconidiation with concomitant increase in brlA and wetA expression levels. Conidiation was also observed even in submerged cultures, a condition that prevented conidiation in the wild type. The effect of rtfA on conidiation also varies in different Aspergillus species. In contrast with A. fumigatus, in A. nidulans and A. flavus the absence of rtfA results in a reduction of conidiation [28, 29]. It is possible that evolutionary rewiring could have occurred resulting in the observed differential regulatory output of rtfA in morphological development in different Aspergillus species.
The conidia that are able to evade the defenses of the lung environment will germinate and grow as hyphae. In our study, the deletion of rtfA lead to colonies that were significantly reduced in vegetative growth (37% decrease) compared to the wild type strain, which may affect the ability of the fungus to establish an infection in the host. These results coincide with previous findings from our lab, where rtfA in A. nidulans and A. flavus was also shown to affect growth [28, 29], although the effect of the deletion of rtfA in A. fumigatus growth was slightly more severe compared to that of A. nidulans and A. flavus.
The human immune system has an array of ways to defend the organism against pathogens such as A. fumigatus. Macrophages and neutrophils produce reactive oxygen species (ROS) that can be detrimental to the fungal spores and hyphae [48]. For this reason, we tested the oxidative stress response of A. fumigatus wild type, ΔrtfA, complementation, and OErtfA strains to various concentrations of menadione. Menadione produces ROS once it enters the cell, creating an environment that is under oxidative stress [73]. Our results conclusively showed that rtfA is important for the response of A. fumigatus to oxidative stress, where the mutant was unable to grow on solid medium in the presence of a 20 μM concentration of menadione, compared to the wild type strain which was still viable at this concentration. Unexpectedly, when gene expression of cat1 and cat2 was assessed in submerged cultures, the ΔrtfA strain presented higher expression compared to the wild type, indicating that rtfA negatively regulates the expression of these genes. Both cat1 and cat2 encode catalases found within the fungal mycelium, in contrast to catA, which is only produced in the conidia [74]. Deletion of cat1 or cat2 does not attenuate virulence of A. fumigatus in vivo, however a double deletion mutant did increase survival [49]. Despite this, it is difficult to assess the inclusion of catalases as virulence factors due to redundancy; the fungus possesses many genes that aid in combating ROS, including five catalases (catA, cat1/catB, catC, catE, and cat2/katG) and four superoxide dismutatases: sod1, sod2, sod3, and sod4 [49, 74, 75]. It is possible that although cat1 and cat2 are expressed at higher levels in the mutant strain, rtfA could be required for other factors involved in oxidative stress resistance.
Although A. fumigatus is known as an important opportunistic human pathogen, this fungus predominantly lives as a saprophytic organism, being found on dead and decaying organic matter. In such an environment, it is important for this organism to produce a plethora of hydrolases in order to obtain nutrients [76]. A majority of these degradative enzymes are specific for plant cell wall components; nevertheless, this fungus does possess some enzymes that can target human tissue, including proteases such as elastase [48, 77]. This is important during lung infection, a microenvironment rich in elastin. In our study we found that the rtfA mutant strain presented a reduction in protease activity with respect to the wild type, indicating the importance of rtfA on nutrient acquisition, and possibly contributing to A. fumigatus pathogenicity.
Aspergillus fumigatus is also known to produce a number of secondary metabolites with a vast spectrum of biological activities. These include a family of ergot alkaloids and the peptidyl alkaloids fumiquinazoline C and tryptoquivaline F, which have been experimentally shown to be associated with conidia and thus dependent on brlA [35, 52, 78, 79]. Though the effect of overexpressing brlA was not examined in that case, it is interesting to note that in our studies the rtfA deletion mutant presented an increase in brlA expression and conidia along with an increase in the production of the ergot alkaloids festuclavine and fumigaclavines A, B, and C, as well as the increase in the peptidyl alkaloid fumiquinazoline C and tryptoquivaline F production. The significance of these classes of alkaloids is revealed in the possible therapeutic applications due to their cytotoxic, anti-inflammatory, anti-tumor, and anti-cancer activities [80, 81]. Another secondary metabolite produced by Aspergillus sp., pseurotin A, has also demonstrated anti-tumor activity as well as anti-parastic activity [82]. Our current study shows that rtfA is a positive regulator of pseurotin A production in A. fumigatus.
The absence of rtfA did not lead to major effects on the integrity of the cell wall, as suggested by the presence of fungal growth when the rtfA mutant was exposed to chemical agents such as SDS. However, our studies indicate that this gene is necessary for normal adhesion capacity to surfaces. This adhesiveness is necessary for the formation of biofilm [83], which also forms in vivo when the fungus is infecting humans [61]. The significance of biofilm is seen in patients infected with a fungal pathogen, where it has been demonstrated in both A. fumigatus as well as Candida albicans to decrease antifungal drug susceptibility [60, 84, 85].
The fact that absence of rtfA results in pleiotropic effects such as alterations in vegetative growth and morphogenesis, resistance to oxidative stress, protease activity, as well as proper adhesion capacity in A. fumigatus could contribute to variations in virulence in this mutant. Indeed, our in vivo analysis of the effect of rtfA on pathogenicity in the larvae of the greater wax moth, Galleria mellonella, revealed a role of rtfA in virulence, as the larvae infected with the deletion mutant showed a significant increase in survival rate compared with those infected with A. fumigatus wild type. Galleria mellonella is often used as a preliminary model system to test virulence due to its cost effectiveness, feasibility, and possession of an innate immune system [86–88]. With these results in mind, we tested an additional in vivo model using neutropenic murine animals. In this second model, the ΔrtfA strain also showed a reduction in virulence compared to that of the wild type, indicating that rtfA is indispensable for establishing normal infections.
In conclusion, this study demonstrates the involvement and importance of the rtfA gene in A. fumigatus. Fundamental biological processes, such as growth and morphological development are regulated by rtfA. We also demonstrated the influence of rtfA on the synthesis of various secondary metabolites as well as protease activity, an important element in nutrient acquisition that could also influence pathogenicity. Other factors also associated with virulence, such as oxidative stress and adhesion capacity were affected by rtfA as well. The profound effect of rtfA in A. fumigatus biology makes this gene indispensable for normal pathogenicity. Indeed, possibly the most relevant finding was the significant decrease in virulence in the absence of rtfA, shown in both the Galleria and murine infection models. This reduction in virulence, together with the fact that A. fumigatus RtfA presents low similarity with homologs of higher eukaryotes, suggest that this regulator could be used as a potential desirable target in the design of a control strategy against Aspergillosis.
Supporting information
(DOCX)
(DOCX)
(A) Schematic diagram showing the replacement of rtfA with the A. parasiticus pyrG gene by a double-crossover event. Southern blot analysis confirming the proper integration of the cassette is shown. The 5’ UTR was used as the probe. SalI (S) was used to digest genomic DNA. Bands of 5.1 kb and 3.7 kb indicate deletion of rtfA in the mutant (ΔrtfA), whereas bands of 5.1 and 1.6 correspond to the wild type (WT, CEA10). The deletion strain was denominated TRRM4. (B) Confirmation of the complementation strain by PCR using primers AfumRM3_Oef (F) and AfumRM3_Oer (R) (S1 Table). The expected PCR product size in the complementation strain is 2036 bp. Wild type (CEA10) genomic DNA was used as a positive control and ΔrtfA DNA was used as negative control. (C) Generation of the over-expression strain was confirmed by PCR analysis, using primers gpdApromoF (F) and AfumRM3_oer (R). The expected product size is 2178 bp. The plasmid used for transformation (pRRM2) was used as positive control. (D) qRT-PCR expression analysis of rtfA in the four strains obtained. The results were normalized to the WT considered as 1. Bars represent standard error. Asterisk: not detected.
(PDF)
Wild type (WT), deletion (ΔrtfA), complementation (com), and overexpression (OErtfA) strains were point-inoculated on GMM supplemented with increasing concentrations of 6-Azauracil. Strains were incubated at 37°C. (A) Photos correspond to 48 h and 72 h cultures (upper and lower rows respectively). Colony diameters were measured at 48 h (B) and 72 h (C). Error bars represent standard error.
(PDF)
(A) Quantification of conidial production on 1% GMM agar plates. Strains were incubated at 37°C for six days and core samples were obtained 0.5 cm from the colony center. Different letters above the bars indicate significantly different values (p ≤ 0.05). Error bars represent standard error. (B) Cultures of ΔrtfA grown in submerged cultures (GMM) accumulated a purple pigment not present in the other strains.
(PDF)
(A) Diagram of the strategy to fuse gfp to rtfA. The transformation cassette contained 885 bp of the rtfA 3’ region. The tagged construct was introduced at the rtfA locus by a double-recombination event as indicated, resulting in the TRRM6 strain. Transformants were verified by diagnostic PCR with primers AfumRM3_Oef (F) and AfumRM33Nested (R) (S1 Table). The expected PCR products for the gfp-tagged strain (5.9 kb) and wild type (3.2 kb) were obtained (right) (B) Micrographs showing the subcellular localization of RtfA::GFP. From left to right, DIC images, DAPI images indicating the position of nuclei, and green fluorescence (GF) images. Arrows indicate nuclei. Scale bars represent 10 μm.
(PDF)
The wild type (WT) and overexpression (OErtfA) strains were tested in the presence of a range of menadione concentrations from 0 to 35 μM. Strains were incubated at 37°C. Photograph was taken after 48 h.
(PDF)
Aspergillus fumigatus wild type (WT), ΔrtfA, complementation (com), and overexpression rtfA (OErtfA) strains were point-inoculated on GMM with different pH values and incubated at 37°C. Colony diameters were measured at 48 h (A) and 72 h (B). Images of the plates were taken at 72 h (C).
(PDF)
Aspergillus fumigatus wild type (WT), ΔrtfA, complementation (com), and overexpression rtfA (OErtfA) strains were point-inoculated on GMM and incubated at a range of temperatures as shown. Colony diameter (A) and images of the plates (B) were obtained at 48 h.
(PDF)
Wild type (WT), deletion (ΔrtfA), complementation (com), and overexpression (OErtfA) strains were point-inoculated onto GMM medium supplemented with 1.2 M sorbitol, 0.6 M KCl, or 1.0 M sucrose to induce osmotic stress. Plates were incubated at 37°C and colony diameters were measured at 48 h (A). Error bars represent standard error. (B) Photographs of colonies from 48 h of incubation.
(PDF)
Wild type (WT), deletion (ΔrtfA), complementation (com), and overexpression (OErtfA) strains were point-inoculated on GMM supplemented with increasing concentrations of SDS. Strains were incubated at 37°C. (A) Photographs of colony at 48 h and 72 h. (B) Colony diameters measured at 48 h and 72 h after inoculation. Error bars represent standard error.
(PDF)
Strains were inoculated in liquid GMM and incubated at 37°C for 24 h (A), 48 h (B), and 72 h (C). After staining with Crystal Violet the absorbance was read at 560 nm. Samples were diluted 3 fold. Different letters indicate statistically different values (p ≤ 0.05). Error bars represent standard error.
(PDF)
Acknowledgments
This project was funded by Northern Illinois University. The authors thank Sourabh Dhingra, Corinna Kashuba, Scott Grayburn, and Xue-Huan Feng for their technical support.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Internal funding from Northern Illinois University supported this research. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Schmitt HJ, Blevins A, Sobeck K, Armstrong D. Aspergillus species from hospital air and from patients. Mycoses. 1990. Nov-Dec; 33: 539–41. [DOI] [PubMed] [Google Scholar]
- 2.Rüchel R, Reichard U. Pathogenesis and clinical presentation of aspergillosis. Contrib Microbiol. 1999; 2: 21–43. [DOI] [PubMed] [Google Scholar]
- 3.Samson RA. The genus Aspergillus with special regard to the Aspergillus fumigatus group. Contrib Microbiol. 1999; 2: 5–20. [DOI] [PubMed] [Google Scholar]
- 4.Hedayati MT, Pasqualotto AC, Warn PA, Bowyer P, Denning DW. Aspergillus flavus: human pathogen, allergen and mycotoxin producer. Microbiology. 2007. June; 153: 1677–92. 10.1099/mic.0.2007/007641-0 [DOI] [PubMed] [Google Scholar]
- 5.Denning DW. Invasive aspergillosis. Clin Infect Dis. 1998. April; 26: 781–803. [DOI] [PubMed] [Google Scholar]
- 6.Kliasova GA, Petrova NA, Parovichnikova EN, Gotman LN, Isaev VG, Mikhalova EA, et al. Invasive pulmonary aspergillosis. Terapevticheskii Arkhiv 2005; 77: 65–71. [PubMed] [Google Scholar]
- 7.Marr KA, Carter RA, Boeckh M, Martin P, Corey L. Invasive aspergillosis in allogeneic stem cell transplant recipients: changes in epidemiology and risk factors. Blood. 2002. December 15; 100: 4358–66. 10.1182/blood-2002-05-1496 [DOI] [PubMed] [Google Scholar]
- 8.Pagano L, Girmenia C, Mele L, Ricci P, Tosti ME, Nosari A, et al. Infections caused by filamentous fungi in patients with hematologic malignancies. A report of 391 cases by GIMEMA Infection Program. Haematologica. 2001. August; 86: 862–70. [PubMed] [Google Scholar]
- 9.Post MJ, Lass-Floerl C, Gastl G, Nachbaur D. Invasive fungal infections in allogeneic and autologous stem cell transplant recipients: a single-center study of 166 transplanted patients. Transpl Infect Dis. 2007. September;9:189–95. 10.1111/j.1399-3062.2007.00219.x [DOI] [PubMed] [Google Scholar]
- 10.Wiederhold NP, Lewis RE, Kontoyiannis DP. Invasive aspergillosis in patients with hematologic malignancies. Pharmacotherapy. 2003. December;23:1592–610. [DOI] [PubMed] [Google Scholar]
- 11.Latgé JP. Aspergillus fumigatus and aspergillosis. Clin Microbiol Rev. 1999. April;12:310–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knutsen AP, Bellone C, Kauffman H. Immunopathogenesis of allergic bronchopulmonary aspergillosis in cystic fibrosis. J Cyst Fibros. 2002. June;1:76–89. [DOI] [PubMed] [Google Scholar]
- 13.Stevens DA, Moss RB, Kurup VP, Knutsen AP, Greenberger P, Judson MA, et al. Allergic bronchopulmonary aspergillosis in cystic fibrosis—state of the art: Cystic Fibrosis Foundation Consensus Conference. Clin Infect Dis. 2003. October 1;37 Suppl 3:S225–64. [DOI] [PubMed] [Google Scholar]
- 14.Kurup VP, Knutsen AP. Allergic bronchopulmonary aspergillosis. Allergy Fron: Clin Manifest. 2009;3:351–66. [Google Scholar]
- 15.Patterson R, Greenberger PA, Radin RC, Roberts M. Allergic bronchopulmonary aspergillosis: staging as an aid to management. Ann Intern Med. 1982. March;96:286–91. [DOI] [PubMed] [Google Scholar]
- 16.Rosenberg M, Patterson R, Mintzer R, Cooper BJ, Roberts M, Harris KE. Clinical and immunologic criteria for the diagnosis of allergic bronchopulmonary aspergillosis. Ann Intern Med. 1977. April;86:405–14. [DOI] [PubMed] [Google Scholar]
- 17.Kontoyiannis DP, Bodey GP. Invasive aspergillosis in 2002: an update. Eur J Clin Microbiol Infect Dis. 2002. March;21:161–72. 10.1007/s10096-002-0699-z [DOI] [PubMed] [Google Scholar]
- 18.Oren I, Goldstein N. Invasive pulmonary aspergillosis. Curr Opin Pulm Med. 2002;8:195–200. [DOI] [PubMed] [Google Scholar]
- 19.Sherif R, Segal BH. Pulmonary aspergillosis: clinical presentation, diagnostic tests, management and complications. Curr Opin Pulm Med. 2010. May;16:242–50. 10.1097/MCP.0b013e328337d6de [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen J, Yang Q, Huang J, Li L. Risk factors for invasive pulmonary aspergillosis and hospital mortality in acute-on-chronic liver failure patients: a retrospective-cohort study. Int J Med Sci. 2013. September 18;10:1625–31. 10.7150/ijms.6824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dhingra S, Andes D, Calvo AM. VeA regulates conidiation, gliotoxin production, and protease activity in the opportunistic human pathogen Aspergillus fumigatus. Eukaryot Cell. 2012. December;11:1531–43. 10.1128/EC.00222-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stolinski LA, Eisenmann DM, Arndt KM. Identification of RTF1, a novel gene important for TATA site selection by TATA box-binding protein in Saccharomyces cerevisiae. Mol Cell Biol. 1997. August; 17:4490–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ng HH, Dole S, Struhl K. The Rtf1 component of the Paf1 transcriptional elongation complex is required for ubiquitination of histone H2B. J Biol Chem. 2003. September 5;278:33625–28. 10.1074/jbc.C300270200 [DOI] [PubMed] [Google Scholar]
- 24.Warner MH, Roinick KL, Arndt KM. Rtf1 is a multifunctional component of the Paf1 complex that regulates gene expression by directing cotranscriptional histone modification. Mol Cell Biol. 2007. September; 27:6103–15. 10.1128/MCB.00772-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Jong RN, Truffault V, Diercks T, Ab E, Daniels MA, Kaptein R, et al. Structure and DNA binding of the human Rtf1 Plus3 domain. Structure. 2008. January;16:149–59. 10.1016/j.str.2007.10.018 [DOI] [PubMed] [Google Scholar]
- 26.Akanuma T, Koshida S, Kawamura A, Kishimoto Y, Takada S. Paf1 complex homologues are required for Notch-regulated transcription during somite segmentation. EMBO Rep. 2007. September;8:858–63. 10.1038/sj.embor.7401045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ding L, Paszkowski-Rogacz M, Nitzsche A, Slabicki MM, Heninger AK, de Vries I, et al. A genome-scale RNAi screen for Oct4 modulators defines a role of the Paf1 complex for embryonic stem cell identity. Cell Stem Cell. 2009. May 8;4:403–15. 10.1016/j.stem.2009.03.009 [DOI] [PubMed] [Google Scholar]
- 28.Ramamoorthy V, Shantappa S, Dhingra S, Calvo AM. veA-dependent RNA-pol II transcription elongation factor-like protein, RtfA, is associated with secondary metabolism and morphological development in Aspergillus nidulans. Mol Microbiol. 2012. August;85:795–814. 10.1111/j.1365-2958.2012.08142.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lohmar JM, Harris-Coward PY, Cary JW, Dhingra S, Calvo AM. rtfA, a putative RNA-Pol II transcription elongation factor gene, is necessary for normal morphological and chemical development in Aspergillus flavus. Appl Microbiol Biotechnol. 2016. June;100:5029–41. 10.1007/s00253-016-7418-7 [DOI] [PubMed] [Google Scholar]
- 30.Szewczyk E, Nayak T, Oakley CE, Edgerton H, Xiong Y, Taheri-Talesh N, et al. Fusion PCR and gene targeting in Aspergillus nidulans. Nat Protoc. 2006;1:3111–20. 10.1038/nprot.2006.405 [DOI] [PubMed] [Google Scholar]
- 31.Smith TD, Calvo AM. The mtfA transcription factor gene controls morphogenesis, gliotoxin production, and virulence in the opportunistic human pathogen Aspergillus fumigatus. Eukaryot Cell. 2014. June;13:766–75. 10.1128/EC.00075-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peñalva MA. Tracing the endocytic pathway of Aspergillus nidulans with FM4-64. Fungal Genet Biol. 2005. December;42:963–75. 10.1016/j.fgb.2005.09.004 [DOI] [PubMed] [Google Scholar]
- 33.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001. December;25:402–08. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 34.Reichard U, Monod M, Odds F, Rüchel R. Virulence of an aspergillopepsin-deficient mutant of Aspergillus fumigatus and evidence for another aspartic proteinase linked to the fungal cell wall. J Med Vet Mycol. 1997. May-Jun;35:189–96. [DOI] [PubMed] [Google Scholar]
- 35.Gauthier T, Wang X, Sifuentes Dos Santos J, Fysikopoulos A, Tadrist S, Canlet C, et al. Trypacidin, a spore-borne toxin from Aspergillus fumigatus, is cytotoxic to lung cells. PLoS One. 2012;7:e29906 10.1371/journal.pone.0029906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cano PM, Jamin EL, Tadrist S, Bourdaud'hui P, Péan M, Debrauwer L, et al. New untargeted metabolic profiling combining mass spectrometry and isotopic labeling: application on Aspergillus fumigatus grown on wheat. Anal Chem. 2013. September 3;85:8412–20. 10.1021/ac401872f [DOI] [PubMed] [Google Scholar]
- 37.Fuchs BB, O'Brien E, Khoury JB, Mylonakis E. Methods for using Galleria mellonella as a model host to study fungal pathogenesis. Virulence. 2010. Nov-Dec;1:475–82. [DOI] [PubMed] [Google Scholar]
- 38.Exinger F, Lacroute F. 6-Azauracil inhibition of GTP biosynthesis in Saccharomyces cerevisiae. Curr Genet. 1992. July;22:9–11. [DOI] [PubMed] [Google Scholar]
- 39.Archambault J, Lacroute F, Ruet A, Friesen JD. Genetic interaction between transcription elongation factor TFIIS and RNA polymerase II. Mol Cell Biol. 1992. September;12:4142–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Costa PJ, Arndt KM. Synthetic lethal interactions suggest a role for the Saccharomyces cerevisiae Rtf1 protein in transcription elongation. Genetics. 2000. October;156:535–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adams TH, Boylan MT, Timberlake WE. brlA is necessary and sufficient to direct conidiophore development in Aspergillus nidulans. Cell. 1988. July 29;54:353–62. [DOI] [PubMed] [Google Scholar]
- 42.Son H, Kim MG, Min K, Lim JY, Choi GJ, Kim JC, et al. WetA is required for conidiogenesis and conidium maturation in the ascomycete fungus Fusarium graminearum. Eukaryot Cell. 2014. January;13:87–98. 10.1128/EC.00220-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kolattukudy PE, Lee JD, Rogers LM, Zimmerman P, Ceselski S, Fox B, et al. Evidence for possible involvement of an elastolytic serine protease in aspergillosis. Infect Immun. 1993. June;61:2357–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Richie DL, Hartl L, Aimanianda V, Winters MS, Fuller KK, Miley MD, et al. A role for the unfolded protein response (UPR) in virulence and antifungal susceptibility in Aspergillus fumigatus. PLoS Pathog. 2009. January;5:e1000258 10.1371/journal.ppat.1000258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jaton-Ogay K, Paris S, Huerre M, Quadroni M, Falchetto R, Togni G, et al. Cloning and disruption of the gene encoding an extracellular metalloprotease of Aspergillus fumigatus. Mol Microbiol. 1994. December; 14:917–28. [DOI] [PubMed] [Google Scholar]
- 46.Bergmann A, Hartmann T, Cairns T, Bignell EM, Krappmann S. A regulator of Aspergillus fumigatus extracellular proteolytic activity is dispensable for virulence. Infect Immun. 2009. September;77:4041–50. 10.1128/IAI.00425-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Latgé JP. The pathobiology of Aspergillus fumigatus. Trends Microbiol. 2001. August;9:382–9. [DOI] [PubMed] [Google Scholar]
- 48.Dagenais TR, Keller NP. Pathogenesis of Aspergillus fumigatus in Invasive Aspergillosis. Clin Microbiol Rev. 2009. July;22:447–65. 10.1128/CMR.00055-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shibuya K, Paris S, Ando T, Nakayama H, Hatori T, Latgé JP. Catalases of Aspergillus fumigatus and inflammation in aspergillosis. Nihon Ishinkin Gakkai Zasshi. 2006;47:249–55. [DOI] [PubMed] [Google Scholar]
- 50.Lind AL, Smith TD, Saterlee T, Calvo AM, Rokas A. Regulation of Secondary Metabolism by the Velvet Complex Is Temperature-Responsive in Aspergillus. G3 (Bethesda). 2016. December 7;6:4023–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wallwey C, Matuschek M, Li SM. Ergot alkaloid biosynthesis in Aspergillus fumigatus: conversion of chanoclavine-I to chanoclavine-I aldehyde catalyzed by a short-chain alcohol dehydrogenase FgaDH. Arch Microbiol. 2010. February;192:127–34. 10.1007/s00203-009-0536-1 [DOI] [PubMed] [Google Scholar]
- 52.Lim FY, Ames B, Walsh CT, Keller NP. Co-ordination between BrlA regulation and secretion of the oxidoreductase FmqD directs selective accumulation of fumiquinazoline C to conidial tissues in Aspergillus fumigatus. Cell Microbiol. 2014. August;16:1267–83. 10.1111/cmi.12284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maiya S, Grundmann A, Li X, Li SM, Turner G. Identification of a hybrid PKS/NRPS required for pseurotin A biosynthesis in the human pathogen Aspergillus fumigatus. Chembiochem. 2007. September 24; 8:1736–43. 10.1002/cbic.200700202 [DOI] [PubMed] [Google Scholar]
- 54.Damveld RA, vanKuyk PA, Arentshorst M, Klis FM, van den Hondel CA, Ram AF. Expression of agsA, one of five 1,3-alpha-D-glucan synthase-encoding genes in Aspergillus niger, is induced in response to cell wall stress. Fungal Genet Biol. 2005. February;42:165–77. 10.1016/j.fgb.2004.11.006 [DOI] [PubMed] [Google Scholar]
- 55.de Groot PW, Ram AF, Klis FM. Features and functions of covalently linked proteins in fungal cell walls. Fungal Genet Biol. 2005. August;42:657–75. 10.1016/j.fgb.2005.04.002 [DOI] [PubMed] [Google Scholar]
- 56.Hill TW, Loprete DM, Momany M, Ha Y, Harsch LM, Livesay JA, et al. Isolation of cell wall mutants in Aspergillus nidulans by screening for hypersensitivity to Calcofluor White. Mycologia. 2006. May-Jun;98:399–409. [DOI] [PubMed] [Google Scholar]
- 57.Pancaldi S, Poli F, Dall'Olio G, Vannini GL. Morphological anomalies induced by Congo red in Aspergillus niger. Arch Microbiol. 1984. March;137:185–87. [DOI] [PubMed] [Google Scholar]
- 58.Roncero C, Durán A. Effect of Calcofluor white and Congo red on fungal cell wall morphogenesis: in vivo activation of chitin polymerization. J Bacteriol. 1985. September;163:1180–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wood PJ. Specificity in the interaction of direct dyes with polysaccharides. Carbohydr Res 1980;85: 271–87. [Google Scholar]
- 60.Wuren T, Toyotome T, Yamaguchi M, Takahashi-Nakaguchi A, Muraosa Y, Yahiro M, Wang DN, Watanabe A, Taguchi H, Kamei K. Effect of serum components on biofilm formation by Aspergillus fumigatus and other Aspergillus species. Jpn J Infect Dis. 2014;67:172–79. [DOI] [PubMed] [Google Scholar]
- 61.Loussert C, Schmitt C, Prevost MC, Balloy V, Fadel E, Philippe B, et al. In vivo biofilm composition of Aspergillus fumigatus. Cell Microbiol. 2010. March;12:405–10. 10.1111/j.1462-5822.2009.01409.x [DOI] [PubMed] [Google Scholar]
- 62.Desai JV, Mitchell AP, Andes DR. Fungal biofilms, drug resistance, and recurrent infection. Cold Spring Harb Perspect Med. 2014. October 1;4 pii:a019729 10.1101/cshperspect.a019729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010. September;8:623–33. 10.1038/nrmicro2415 [DOI] [PubMed] [Google Scholar]
- 64.Kelleher RJ 3rd, Flanagan PM, Kornberg RD. A novel mediator between activator proteins and the RNA polymerase II transcription apparatus. Cell. 1990. June 29;61:1209–15. [DOI] [PubMed] [Google Scholar]
- 65.Kim M, Ahn SH, Krogan NJ, Greenblatt JF, Buratowski S. Transitions in RNA polymerase II elongation complexes at the 3' ends of genes. EMBO J. 2004. January 28;23:354–64. 10.1038/sj.emboj.7600053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mayer A, Lidschreiber M, Siebert M, Leike K, Söding J, Cramer P. Uniform transitions of the general RNA polymerase II transcription complex. Nat Struct Mol Biol. 2010. October;17:1272–78. 10.1038/nsmb.1903 [DOI] [PubMed] [Google Scholar]
- 67.Briggs SD, Xiao T, Sun ZW, Caldwell JA, Shabanowitz J, Hunt DF, et al. Gene silencing: trans-histone regulatory pathway in chromatin. Nature. 2002. August 1;418:498 10.1038/nature00970 [DOI] [PubMed] [Google Scholar]
- 68.Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002. July 4;418:104–48. 10.1038/nature00883 [DOI] [PubMed] [Google Scholar]
- 69.Dover J, Schneider J, Tawiah-Boateng MA, Wood A, Dean K, Johnston M, et al. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J Biol Chem. 2002. August 9; 277:28368–71. 10.1074/jbc.C200348200 [DOI] [PubMed] [Google Scholar]
- 70.Ng HH, Xu RM, Zhang Y, Struhl K. Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J Biol Chem. 2002. September 20;277:34655–57. 10.1074/jbc.C200433200 [DOI] [PubMed] [Google Scholar]
- 71.Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, et al. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol Cell. 2003. March;11:721–29. [DOI] [PubMed] [Google Scholar]
- 72.Wind M, Reines D. Transcription elongation factor SII. Bioessays. 2000. April;22:327–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.de Sá RA, de Castro FA, Eleutherio EC, de Souza RM, da Silva JF, Pereira MD. Brazilian propolis protects Saccharomyces cerevisiae cells against oxidative stress. Braz J Microbiol. 2013. December 17;44:993–1000. 10.1590/S1517-83822013005000062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Calera JA, Paris S, Monod M, Hamilton AJ, Debeaupuis JP, Diaquin M, et al. Cloning and disruption of the antigenic catalase gene of Aspergillus fumigatus. Infect Immun. 1997. November;65:4718–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Osherov N. The virulence of Aspergillus fumigatus Kavanagh K, editor. In: New insights in medical mycology. 2007. p. 185–212. [Google Scholar]
- 76.de Vries RP, Visser J. Aspergillus enzymes involved in degradation of plant cell wall polysaccharides. Microbiol Mol Biol Rev. 2001. December;65:497–522. 10.1128/MMBR.65.4.497-522.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kothary MH, Chase T Jr, Macmillan JD. Correlation of elastase production by some strains of Aspergillus fumigatus with ability to cause pulmonary invasive aspergillosis in mice. Infect Immun. 1984. January;43:320–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Panaccione DG, Coyle CM. Abundant respirable ergot alkaloids from the common airborne fungus Aspergillus fumigatus. Appl Environ Microbiol. 2005. June;71:3106–11. 10.1128/AEM.71.6.3106-3111.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Coyle CM, Kenaley SC, Rittenour WR, Panaccione DG. Association of ergot alkaloids with conidiation in Aspergillus fumigatus. Mycologia. 2007. Nov-Dec;99:804–11. [DOI] [PubMed] [Google Scholar]
- 80.Li YX, Himaya SW, Dewapriya P, Zhang C, Kim SK. Fumigaclavine C from a marine-derived fungus Aspergillus fumigatus induces apoptosis in MCF-7 breast cancer cells. Mar Drugs. 2013. December 13; 11:5063–86. 10.3390/md11125063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Han X, Xu X, Cui C, Gu Q. Alkaloidal compounds produced by a marine-derived fungus, Aspergillus fumigatus H1-04, and their antitumor activities. Chin J Med Chem. 2007;17:232–37. [Google Scholar]
- 82.Martínez-Luis S, Cherigo L, Arnold E, Spadafora C, Gerwick WH, Cubilla-Rios L. Antiparasitic and anticancer constituents of the endophytic fungus Aspergillus sp. strain F1544. Nat Prod Commun. 2012. February;7:165–68. [PubMed] [Google Scholar]
- 83.Beauvais A, Schmidt C, Guadagnini S, Roux P, Perret E, Henry C, et al. An extracellular matrix glues together the aerial-grown hyphae of Aspergillus fumigatus. Cell Microbiol. 2007. June;9:1588–600. 10.1111/j.1462-5822.2007.00895.x [DOI] [PubMed] [Google Scholar]
- 84.Seidler MJ, Salvenmoser S, Müller FM. Aspergillus fumigatus forms biofilms with reduced antifungal drug susceptibility on bronchial epithelial cells. Antimicrob Agents Chemother. 2008. November;52:4130–36. 10.1128/AAC.00234-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ramage G, Saville SP, Thomas DP, López-Ribot JL. Candida biofilms: an update. Eukaryot Cell. 2005. April;4:633–38. 10.1128/EC.4.4.633-638.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu Rev Immunol. 2007; 25:697–743. 10.1146/annurev.immunol.25.022106.141615 [DOI] [PubMed] [Google Scholar]
- 87.Fallon J, Kelly J, Kavanagh K. Galleria mellonella as a model for fungal pathogenicity testing. Methods Mol Biol. 2012;845:469–85. 10.1007/978-1-61779-539-8_33 [DOI] [PubMed] [Google Scholar]
- 88.Ramarao N, Nielsen-Leroux C, Lereclus D. The insect Galleria mellonella as a powerful infection model to investigate bacterial pathogenesis. J Vis Exp. 2012. December 11;(70):e4392 10.3791/4392 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(A) Schematic diagram showing the replacement of rtfA with the A. parasiticus pyrG gene by a double-crossover event. Southern blot analysis confirming the proper integration of the cassette is shown. The 5’ UTR was used as the probe. SalI (S) was used to digest genomic DNA. Bands of 5.1 kb and 3.7 kb indicate deletion of rtfA in the mutant (ΔrtfA), whereas bands of 5.1 and 1.6 correspond to the wild type (WT, CEA10). The deletion strain was denominated TRRM4. (B) Confirmation of the complementation strain by PCR using primers AfumRM3_Oef (F) and AfumRM3_Oer (R) (S1 Table). The expected PCR product size in the complementation strain is 2036 bp. Wild type (CEA10) genomic DNA was used as a positive control and ΔrtfA DNA was used as negative control. (C) Generation of the over-expression strain was confirmed by PCR analysis, using primers gpdApromoF (F) and AfumRM3_oer (R). The expected product size is 2178 bp. The plasmid used for transformation (pRRM2) was used as positive control. (D) qRT-PCR expression analysis of rtfA in the four strains obtained. The results were normalized to the WT considered as 1. Bars represent standard error. Asterisk: not detected.
(PDF)
Wild type (WT), deletion (ΔrtfA), complementation (com), and overexpression (OErtfA) strains were point-inoculated on GMM supplemented with increasing concentrations of 6-Azauracil. Strains were incubated at 37°C. (A) Photos correspond to 48 h and 72 h cultures (upper and lower rows respectively). Colony diameters were measured at 48 h (B) and 72 h (C). Error bars represent standard error.
(PDF)
(A) Quantification of conidial production on 1% GMM agar plates. Strains were incubated at 37°C for six days and core samples were obtained 0.5 cm from the colony center. Different letters above the bars indicate significantly different values (p ≤ 0.05). Error bars represent standard error. (B) Cultures of ΔrtfA grown in submerged cultures (GMM) accumulated a purple pigment not present in the other strains.
(PDF)
(A) Diagram of the strategy to fuse gfp to rtfA. The transformation cassette contained 885 bp of the rtfA 3’ region. The tagged construct was introduced at the rtfA locus by a double-recombination event as indicated, resulting in the TRRM6 strain. Transformants were verified by diagnostic PCR with primers AfumRM3_Oef (F) and AfumRM33Nested (R) (S1 Table). The expected PCR products for the gfp-tagged strain (5.9 kb) and wild type (3.2 kb) were obtained (right) (B) Micrographs showing the subcellular localization of RtfA::GFP. From left to right, DIC images, DAPI images indicating the position of nuclei, and green fluorescence (GF) images. Arrows indicate nuclei. Scale bars represent 10 μm.
(PDF)
The wild type (WT) and overexpression (OErtfA) strains were tested in the presence of a range of menadione concentrations from 0 to 35 μM. Strains were incubated at 37°C. Photograph was taken after 48 h.
(PDF)
Aspergillus fumigatus wild type (WT), ΔrtfA, complementation (com), and overexpression rtfA (OErtfA) strains were point-inoculated on GMM with different pH values and incubated at 37°C. Colony diameters were measured at 48 h (A) and 72 h (B). Images of the plates were taken at 72 h (C).
(PDF)
Aspergillus fumigatus wild type (WT), ΔrtfA, complementation (com), and overexpression rtfA (OErtfA) strains were point-inoculated on GMM and incubated at a range of temperatures as shown. Colony diameter (A) and images of the plates (B) were obtained at 48 h.
(PDF)
Wild type (WT), deletion (ΔrtfA), complementation (com), and overexpression (OErtfA) strains were point-inoculated onto GMM medium supplemented with 1.2 M sorbitol, 0.6 M KCl, or 1.0 M sucrose to induce osmotic stress. Plates were incubated at 37°C and colony diameters were measured at 48 h (A). Error bars represent standard error. (B) Photographs of colonies from 48 h of incubation.
(PDF)
Wild type (WT), deletion (ΔrtfA), complementation (com), and overexpression (OErtfA) strains were point-inoculated on GMM supplemented with increasing concentrations of SDS. Strains were incubated at 37°C. (A) Photographs of colony at 48 h and 72 h. (B) Colony diameters measured at 48 h and 72 h after inoculation. Error bars represent standard error.
(PDF)
Strains were inoculated in liquid GMM and incubated at 37°C for 24 h (A), 48 h (B), and 72 h (C). After staining with Crystal Violet the absorbance was read at 560 nm. Samples were diluted 3 fold. Different letters indicate statistically different values (p ≤ 0.05). Error bars represent standard error.
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.