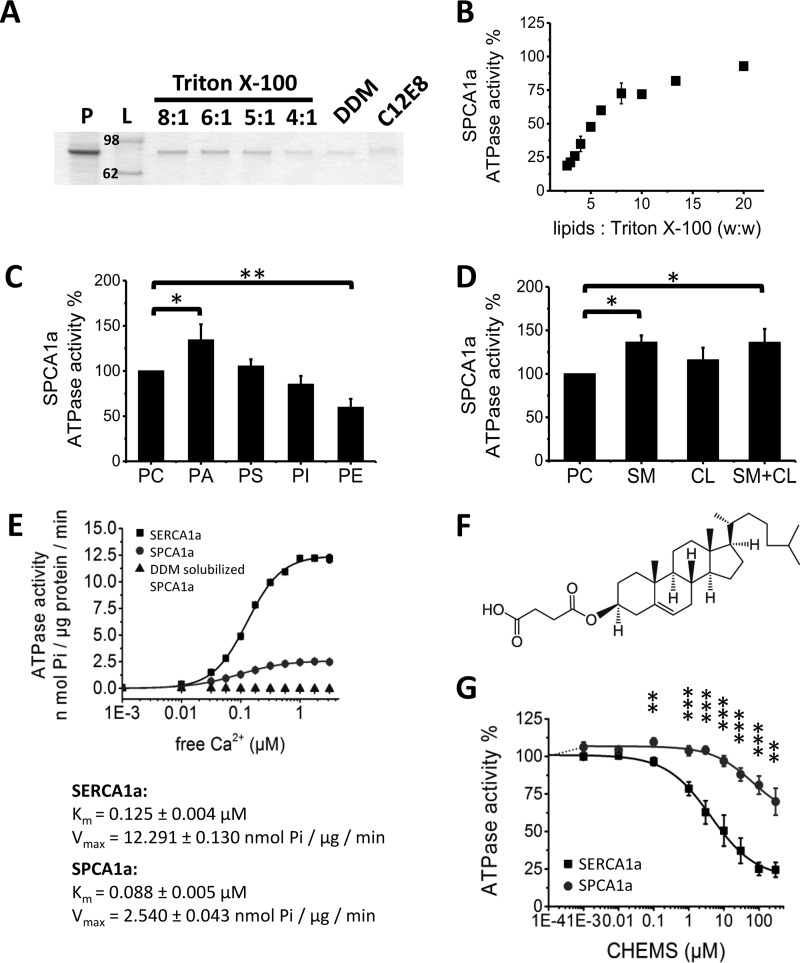
Figure 2.
Optimization of SPCA1a reconstitution into proteoliposomes. A, SDS-polyacrylamide gel stained with SPCA1a proteoliposomes generated with the detergents Triton X-100, DDM, or C12E8. Same volume of samples was loaded on the gel after reconstitution. The lipid/Triton X-100 ratios (w/w) are indicated above the lanes. B, comparison of the SPCA1a ATPase activity following reconstitution with a different (w/w) ratio of lipids/Triton X-100. C and D, 20% weight percent of the indicated lipid(s) were supplemented to PC for SPCA1a reconstitution, and the maximal Ca2+-dependent ATPase activity was determined at 1 μm free Ca2+ concentration (n ≥ 3). The ATPase activities were normalized to the activity when only PC was used. Comparison of the specific SPCA1a ATPase activities in the presence of indicated phospholipids (C), CL, SM, or the combination of CL and SM (1:1 molar ratio) (D). E, comparison of the specific ATPase activities between SERCA1a and SPCA1a in proteoliposomes, and SPCA1a in lipid-free, DDM-solubilized state (n = 3). The DDM-solubilized SPCA1a was purified without supplement of PC in the buffers. F, structure of CHEMS. G, dose-responses of CHEMS on reconstituted SERCA1a and SPCA1a ATPase activities. P, purified SPCA1a; L, ladder; PA, phosphatidic acid; PE, phosphatidylethanolamine; PS, phosphatidylserine; PI, phosphatidylinositol; CL, cholesterol; SM, sphingomyelin; CHEMS, cholesteryl hemisuccinate. *, p < 0.05; **, p < 0.01; ***, p < 0.001.