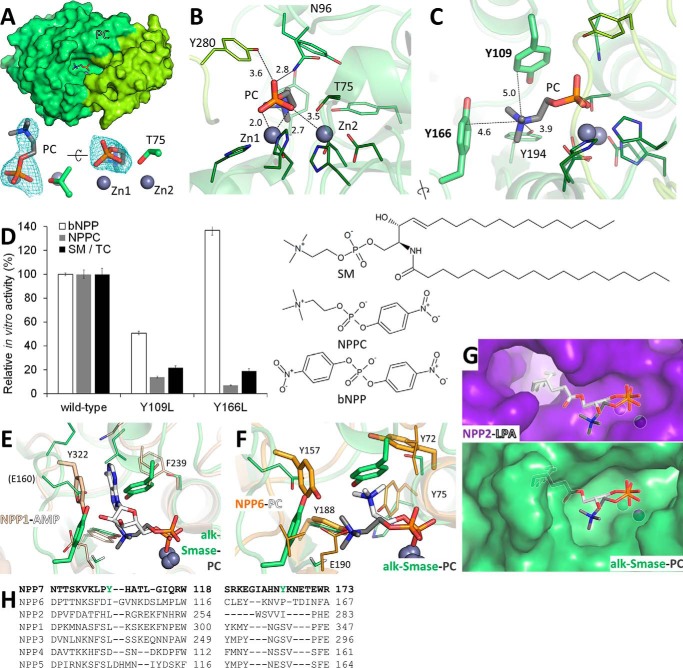
Figure 2.
Recognition of PC by alk-SMase. A, PC (sticks) bound to alk-SMase. Zinc ions (gray spheres) and the catalytic threonine (sticks) are labeled. The electron density Fo − Fc simulated annealing omit map around PC is contoured at 2.5 σ. B, protein residues (sticks) contacting the phosphate group of PC are labeled. C, protein residues (sticks) interacting with the choline moiety of PC are labeled, including the tyrosine side chains forming the aromatic box (bold, thicker sticks). Interatomic distances are indicated (Å). D, enzymatic activity of purified alk-SMase on substrates with chemical structures displayed. 100% activity corresponds to 2.58 and 61.68 μm of p-nitrophenol and 44.72 μm of PC released per nm protein per hour from bNPP, NPPC, and SM in TC micelles, respectively. The values are the means and standard deviations of triplicates representative of one of two experiments. E, superimposition of the PC-alk-SMase complex (gray and green sticks) with the AMP-NPP1 complex (white and beige sticks, PDB code 4GTW). The tyrosine residue of NPP1 stacking with the adenine ring is in thicker sticks. F, superimposition of the PC-alk-SMase complex (gray and green sticks) with the PC-NPP6 complex (white and orange sticks, PDB code 5EGH). For both proteins, the tyrosine side chains forming an aromatic box are in thicker sticks. Key residues of NPP1 and NPP6 are labeled. G, comparison of the substrate binding site of autotaxin (purple, PDB code 3NKN) in complex with lysophosphatidic acid (white sticks) with that of alk-SMase bound to PC (green and gray sticks). Both ligands are superimposed on both proteins. H, structure-assisted sequence alignments of the regions containing the aromatic box of NPP7 (green).