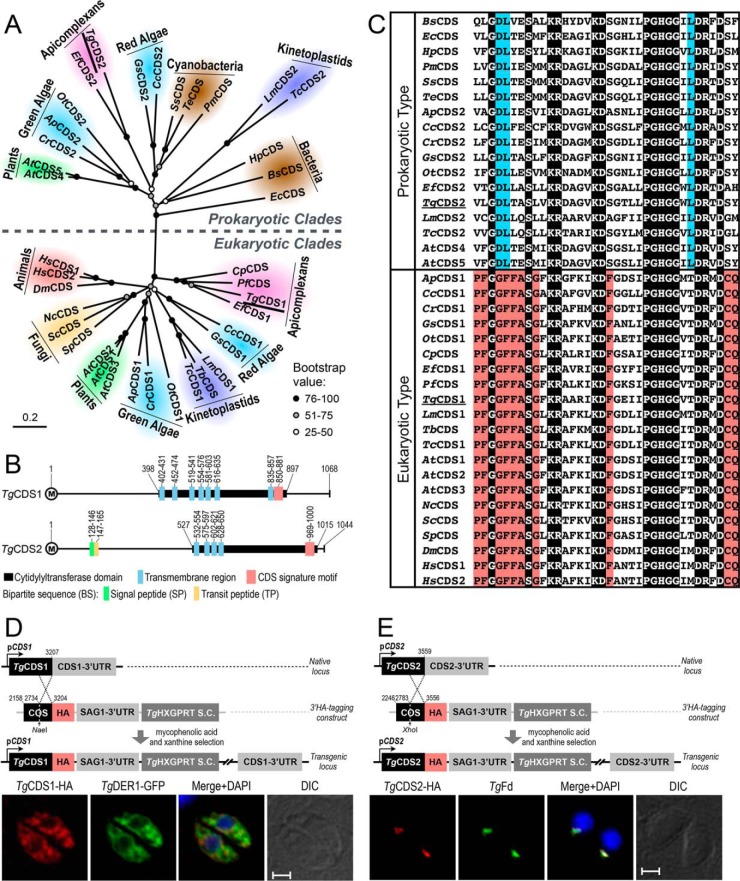
Figure 2.
T. gondii expresses two distinct CDS enzymes located in the ER and apicoplast. A, phylogenetic analysis of TgCDS1, TgCDS2, and orthologs from various organisms representing major forms of life. Branch support was estimated by 100 bootstrap replicates. Other relevant information including the full names of organisms, accession numbers, identity, and similarity details for each CDS sequence are shown in supplemental Table S1. B, schematic drawing of the primary structures of TgCDS1 and TgCDS2. The numbers indicate positions of cytidylyltransferase domains, CDS signature motifs, and transmembrane regions as well as signal and transit peptides, as predicted by Simple Modular Architecture Research Tool (SMART), transmembrane hidden Markov model (TMHMM), SignalP 4.1, ChloroP 1.1, and PlasmoAP. C, multiple alignments of the signature motifs present in eukaryotic- and prokaryotic-type CDS sequences. The residues identical across all sequences are shaded in black, and amino acids that are conserved only in eukaryotes or prokaryotes are highlighted in red or blue, respectively. D and E, scheme for 3′-insertional tagging of TgCDS1 and TgCDS2 genes with a C-terminal HA tag. Plasmids harboring the COS of respective genes were linearized with the indicated enzymes and transfected into tachyzoites (RHΔku80-Δhxgprt) followed by drug selection. Immunofluorescence of stable transgenic tachyzoites expressing TgCDS1-HA (D) and TgCDS2-HA (E) under the control of endogenous promoters and TgSAG1–3′-UTR was performed using anti-HA and Alexa594 antibodies (24 h post-infection). TgCDS1-HA and TgCDS2-HA proteins were co-localized with TgDER1-GFP signal and anti-TgFd/Alexa488 antibodies, respectively. Scale bars: 2 μm. DIC, differential interference contrast.